DNA barcoding snake venoms helps toxinologists save lives
Friday, June 2nd, 2006
 If you hear this sound
If you hear this sound  watch your step–a venomous rattlesnake may be nearby! Prompt administration of the correct antivenom can be life-saving.
watch your step–a venomous rattlesnake may be nearby! Prompt administration of the correct antivenom can be life-saving.
The essential first step in toxinological research is reproducible analyses of venom toxins. However, in December 2005 Toxicon 46: 711-715 Pook and McEwing, University of Wales, report that reproducibility of toxin analyses is commonly compromised by “misidentification of species due to insufficient or changing knowledge of current snake systematics.”
Their article describes a breakthrough in toxinological research: DNA barcoding of dried venoms to provide an accurate, permanent “label” that is independent of future taxonomic changes. This short article is a powerful demonstration of how a standardized approach to identifying species by DNA that uses a public database linked to vouchered specimens, i.e. DNA barcoding, can benefit science and society. While some taxonomists will continue to wring their hands, others may be excited by this work, as it demonstrates how DNA barcoding ADDED TO (and not replacing) standard systematic practice can improve the accessibility and usefulness of ongoing taxonomic work for the larger biological community. I believe this article is enormously important, and I quote here at length.
From the Introduction: “Erroneous or uncertain taxonomy confuses the interpretation of results with respect to intraspecific and interspecific variation. Genuine logistical problems are encountered by non-specialists in taxonomy and snake systematics, in being able to keep up with taxonomic changes. The main hindrances include unresolved taxonomy for some species groups, and new data that change the definition of taxonomic units in others, with the result that the use of systematic information in the toxinological and clinical literature is disorganised. The problem is compounded further by changes in the concept of a particular species, and hence the interpretability of the venom used. Taxonomic confusion, however, has serious implications in snake venom research. The development of effective antivenom treatments and treatment strategies for envenomized patients necessitates a sound taxonomic framework. Accurate clarification of the identity of a given venom is paramount, even after taxonomic revisions involving the species concerned.”
Pook and McEwing “propose a solution to the long-term problem of species identification in toxinological work, utilising mitochondrial haplotypes isolated directly from snake venoms to provide a means of identification that will remain useful even in the face of radical changes in our understanding of the species concerned.”
From the conclusion: “The barcode strategy is dependent on the availablity of known mtDNA haplotype standards against which the sequences being investigated can be compared. Mitochondrial haplotype standards should be gene sequences from snakes of confirmed identity, which have been accessioned to a museum collection as voucher specimens.”


 A year ago in Science,
A year ago in Science, 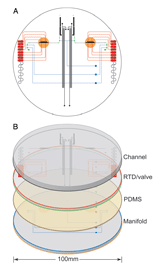 ABI’s smallest sequencer is about the size and weight of a house air conditioning unit [ABI 310: 95 kg (208 lbs); 61 x 56 x 86 cm (24 x 22 x 34 in)]. Researchers who developed the
ABI’s smallest sequencer is about the size and weight of a house air conditioning unit [ABI 310: 95 kg (208 lbs); 61 x 56 x 86 cm (24 x 22 x 34 in)]. Researchers who developed the