RealClear Energy posted an article by Iddo Wernick on consequences of nations choosing different energy trajectories in the Indo-Pacific region in the year 2050. See The Indo-Pacific in 2050: Alternative Energy Scenarios and Security
Area of Research: Oceans
News from the Quiet Ocean Experiment
Terry Collins artfully summarizes the progress in this news release about the International Quiet Ocean Experiment. The news was picked up by
Agencia EFE: Artificial intelligence listens to the habits of marine life (in Spanish)
Independent (London): Scientists eavesdrop on underwater creatures to gain insights on ocean life
Earth.Com: Monitoring ocean life through underwater soundscapes
Portal R7 (Brazil) Biólogos marinhos captam zumbido não identificado que pode ser uma nova espécie de peixe Marine biologists capture unidentified tinnitus that can be a new species of fish
Vice / Motherboard (USA) Scientists Recording Ocean Sounds Picked Up a Mysterious ‘Buzz’ They Can’t Identify
ORF Online (Austria) Unterwassermikrofone belauschen Fische Underwater microphones eavesdrop on fish
Scientias, Netherlands Moet je horen! Vissen maken fascinerende balts- en eetgeluiden, vooral bij volle maan You have to hear! Fish make fascinating balts and eating noises, especially at full moon
Interim Evaluation of International Quiet Ocean Experiment
Jesse co-authored this Interim Program Self-Evaluation of the International Quiet Ocean Experiment for the International Scientific Steering Committee meeting upcoming 26-27 April 2023.
Mark & Jesse present NOAA ‘Omics seminar on eDNA abundance
Mark Stoeckle and Jesse Ausubel presented in the NOAA ‘Omics Seminar Series on Marine fish eDNA Metabarcoding: Promising Developments and Early Applications. The outline:
–eDNA abundance matters (relevant to detection, quantification, field design, laboratory protocols)
–Adding internal standard to metabarcoding PCRs quantifies eDNA (converts relative sequence reads to absolute eDNA copies)
–Current marine fish metabarcoding protocols ready for wider use (reasonably accurate index of fish abundance, especially for more abundant species)
–eDNA metabarcoding can overcome information hurdles for ecosystem-based management
A recording is here , 40-minute presentation and 20-minute Q&A. Thanks to NOAA’s Katharine Egan and Nicole Miller.
Interspecific allometric scaling in eDNA production among northwestern Atlantic bony fishes reflects physiological allometric scaling
Our paper on eDNA as bioassay of Anthropocene published
The new journal based in China, The Innovation, has published the Thaler-Ausubel-Stoeckle paper on Human and domesticated animal environmental DNA as bioassays of the Anthropocene in their “Out of the Box” category, where we like to be. We also post the pdf.
We thank Song Sun and Ke Chen for editorial assistance.
Summary: Human and domesticated animal sequences, commonly detected in environmental DNA (eDNA) metabarcoding studies, are routinely excluded from analysis. Here we suggest that reporting human and domesticated animal eDNA results might open new lines of investigation. For example, the relative abundance of human and domesticated animal eDNA as compared to that of wild vertebrate species might provide an index of human impact on local biota. Such an index could be applied to sites ranging from urban harbors to remote villages, and possibly to analyze historical samples. Various potential sources of contamination complicate the picture, but it should be possible to develop procedures that minimize risk of DNA introduction during collection and processing. Our near-term recommendation is to encourage inclusion of human and domesticated animal data in eDNA publications as an incentive for discovery, to lift quality controls, and to collectively contribute to new vistas that eDNA science might open.
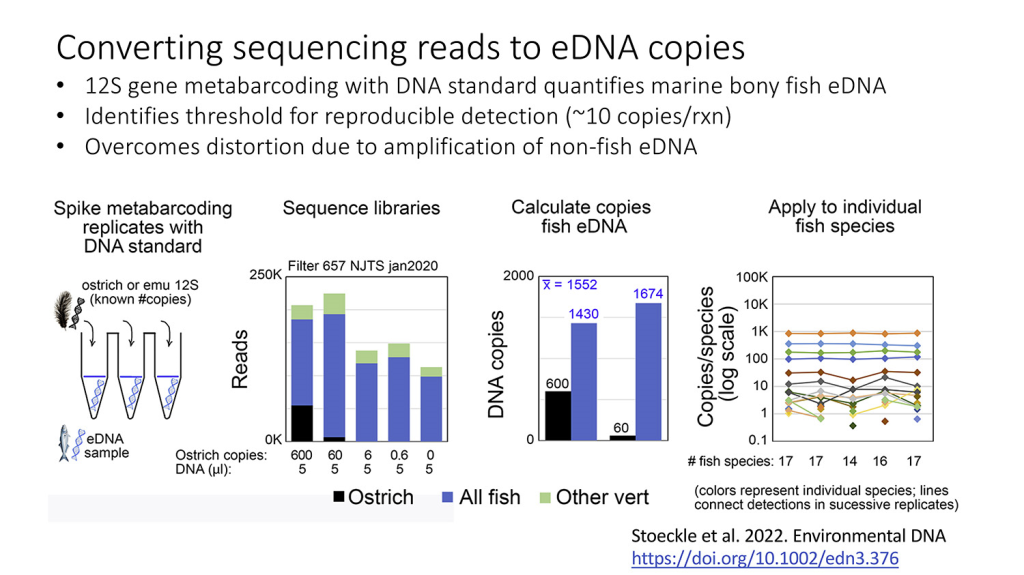
12S gene metabarcoding with DNA standard quantifies marine bony fish environmental DNA, identifies threshold for reproducible detection, and overcomes distortion due to amplification of non-fish DNA
Our article on incorporating a known amount of non-fish DNA to allow better quantification of the fish DNA present in a seawater sample appears in the journal Environmental DNA.
Open Access
12S gene metabarcoding with DNA standard quantifies marine bony fish environmental DNA, identifies threshold for reproducible detection, and overcomes distortion due to amplification of non-fish DNA. Mark Y. Stoeckle, Jesse H. Ausubel, Michael Coogan, first published: 08 December 2022, https://doi.org/10.1002/edn3.376
While our paper focuses on fish, we believe the approach of “spiking” samples collected in nature with known amounts of DNA from a species that would not be present in the sample (such as ostrich) offers great promise for increasing the value of a wide range of aquatic DNA studies. The exhibit below shares some of the main points from the paper.
New code for analyzing eDNA sequences using DADA2 pipeline
For the past year MIchael Coogan, now a grad student in marine science at the U. of New Hampshire, has helped Mark Stoeckle and PHE with improved software for our eDNA studies. See summary below. A pdf of the R code is available here. If you have questions, please write to Mark.
The goal is to adapt the DADA2 pipeline to Mark Stoeckle’s 12S experiment. Sample sequences will be identified using 12S reference file containing sequences of 262 unique vertebrates found around New York. The starting point is a set of Illumina-sequenced paired-end fastq files that have been split (or demultiplexed) by sample and which have barcodes/adapters already removed. The end product will be a sequence table, analogous to the ubiquitous “OTU table”, which records the number of times sample sequences were observed in each sample. The key difference between the output of DADA2 and standard OTU analyses is that DADA2 infers sample sequences exactly rather than clustering sequences into fuzzy OTUs which hide and complicate biological variation.
Fishing for DNA in the Hudson
In the morning of 3 November 2022, Jesse Ausubel summarized the work led by Mark Stoeckle about catching eDNA in the waters of New York City to learn about our local fish species. The site was the beautiful new Little Island in the Hudson River Estuary Park. We post the deck of slides which span from Central Park to the Hudson and East rivers to the Gowanus Canal and Coney Island. New York City enjoys the presence of dozens of fish species, ranging from eels to sturgeon.