Adam Green, of the Economist magazine’s Back to Blue Initiative, interviews Jesse here about industrialization of the oceans.
And refers to our eDNA work in this article on Saving the Bays.
Adam Green, of the Economist magazine’s Back to Blue Initiative, interviews Jesse here about industrialization of the oceans.
And refers to our eDNA work in this article on Saving the Bays.
The International Quiet Ocean Experiment (IQOE) in 2023 gave birth to World Ocean Passive Acoustic Monitoring (WOPAM) Day. The excellent 2024 WOPAM Day video can be seen at https://x.com/GLUBS1/status/1798995048522277190. The full soundtrack for WOPAM Day 2024 is at: https://www.wo-pam.com/wopam-2024 (scroll down and see the link). Congratulations to Miles Parsons and other team members on extending the 2024 monitoring to about 400 sites around the world!
NOAA has posted the video of their ‘Omics Seminar Series: eDNA-Dominant Marine Fish Species Characterize Coastal Habitats presented on 28 February, 2024 by Mark Stoeckle and Jesse Ausubel. The 1-hour seminar is full of new results and ideas about using eDNA data to characterize marine regions and features Mark’s excellent graphics.
Title: eDNA-Dominant Marine Fish Species Characterize Coastal Habitats: an eDNA-Based Classifier Approach to Aid Marine Biogeography and Ocean Monitoring by Mark Stoeckle & Jesse Ausubel
Abstract: A small minority of species typically account for the great majority of individuals or biomass. Here we characterize marine coastal habitats based on abundance of marine fish environmental DNA. We designate the ten most eDNA-abundant fish species in each habitat as eDNA-dominant species. eDNA-dominant species are similar within but differ among habitats and seasons and accord with abundance by traditional survey methods. “Classifiers” based on eDNA-dominant fish species could help map marine fish habitats and monitor changing oceans. Advantages include relatively low sampling requirements, a single technology applicable to diverse habitats, and ease of application to multiple datasets.
RockEDU summer students Priyam Shah and Michael Epelman, who just completed high school, teamed with mentor extraordinaire Mark Stoeckle to study the fishes of an NYC Superfund Site, Newtown Creek. Their excellent poster shows that eDNA detected a surprising diversity of fish in Newtown Creek, despite ongoing pollution and sewage overflow. The number and relative abundance of fish species differed among sites consistent with species habitat preference and pollution tolerance. Our data support eDNA as a cost-effective, non-destructive method for monitoring fish populations and assessing habitat restoration efforts in Newtown Creek and other Superfund sites
A stalwart member of the International Scientific Steering Committee of the Census of Marine Life, Vera Alexander passed away at the age of 90 in Fairbanks AK in May. The Arctic Research Consortium US earlier offered this informative tribute.
Jesse worked closely with Vera during the Census of Marine Life from 1999-2010 and offers this remembrance of The Many Contributions of Vera Alexander.
A child of the International Quiet Ocean Experiment, today is the first World Ocean Passive Acoustics Monitoring (WOPAM) day. The IQOE leaders, including Miles Parsons and Steve Simpson, have prepared two very cool 90-second videos to initiate WOPAM day.
Downloadable at this site. 150 MB each, but they download quickly.
For a 50th Harvard College Reunion Seminar on EO Wilson’s proposal to conserve half Earth, Jesse Ausubel and Mark Stoeckle, assisted by Elizabeth Munnell, conducted a survey of vertebrates in three locations in the Charles River and two in Boston Harbor. The 14 slides on The Charles River and Boston Harbor Then and Now tell a story of remarkable ecological recovery.
Swiss bioinformatics wizard Wandrille Duchemin and PHE Guest Investigator David Thaler publish PyKleeBarcode: Enabling representation of the whole animal kingdom in information space in PLoS One. The computational advances in the paper open the way to calculating DNA-relatedness of all animal species, as the figure below for mammals suggests.
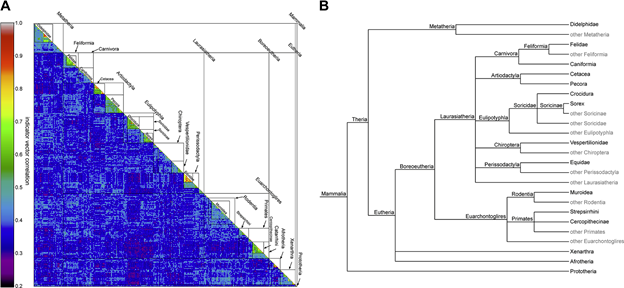
Fig 2. A. View of the structure matrix of the mammalian dataset and taxonomic structure of Mammalia. B. Phylogenetic tree structure of the taxonomic groups retrieved from NCBI taxonomy.
The paper builds on the pioneering work done earlier in the PHE by Larry Sirovich and Mark Stoeckle:
L Sirovich, MY Stoeckle, Y Zhang. A scalable method for analysis and display of DNA sequences. PLoS ONE 4 (10): e7051, 2009
L Sirovich, MY Stoeckle, Y Zhang. Structural analysis of biodiversity. PLoS ONE 5 (2): e9266, 2010
MY Stoeckle, C Coffran. TreeParser-Aided Klee Diagrams Display Taxonomic Clusters in DNA Barcode and Nuclear Gene Datasets . Nature Scientific Reports 3 (2635): 2013
Terry Collins artfully summarizes the progress in this news release about the International Quiet Ocean Experiment. The news was picked up by
Agencia EFE: Artificial intelligence listens to the habits of marine life (in Spanish)
Independent (London): Scientists eavesdrop on underwater creatures to gain insights on ocean life
Earth.Com: Monitoring ocean life through underwater soundscapes
Portal R7 (Brazil) Biólogos marinhos captam zumbido não identificado que pode ser uma nova espécie de peixe Marine biologists capture unidentified tinnitus that can be a new species of fish
Vice / Motherboard (USA) Scientists Recording Ocean Sounds Picked Up a Mysterious ‘Buzz’ They Can’t Identify
ORF Online (Austria) Unterwassermikrofone belauschen Fische Underwater microphones eavesdrop on fish
Scientias, Netherlands Moet je horen! Vissen maken fascinerende balts- en eetgeluiden, vooral bij volle maan You have to hear! Fish make fascinating balts and eating noises, especially at full moon
Mark Stoeckle and Jesse Ausubel presented in the NOAA ‘Omics Seminar Series on Marine fish eDNA Metabarcoding: Promising Developments and Early Applications. The outline:
–eDNA abundance matters (relevant to detection, quantification, field design, laboratory protocols)
–Adding internal standard to metabarcoding PCRs quantifies eDNA (converts relative sequence reads to absolute eDNA copies)
–Current marine fish metabarcoding protocols ready for wider use (reasonably accurate index of fish abundance, especially for more abundant species)
–eDNA metabarcoding can overcome information hurdles for ecosystem-based management
A recording is here , 40-minute presentation and 20-minute Q&A. Thanks to NOAA’s Katharine Egan and Nicole Miller.