The analysis that Jesse Ausubel and Arnulf Gruebler published in 1995 on Working Less and Living Longer: Long-Term Trends in Working Time and Time Budgets is cited in an article on Stressful Leisure in World Magazine, a Christian weekly. We had read the January 2006 report by Mark Aguiar and Eric Hurst on Measuring Trends in Leisure: The Allocation of Time over Five Decades as well as its foil, A Century of Work and Leisure by Valerie Ramey and Neville Francis (June 2005) and recommend both. We continue to believe that changes in total life hours worked merits more attention.
News
Oklahoma Wind Power
The 17 May New York Times quoted Jesse about windpower. For an exposition of the problem, see “Nuclear and Renewable Heresies” posted 19 March 2005.
Salamanders support standardization
In Barcoding Life, Illustrated we suggested that “standardization typically lowers costs and lifts reliability…and should help accelerate construction of comprehensive, consistent reference library of DNA sequences and development of economical technologies for species identification.” However, some have been doubtful about the benefits of standardization. This reflects in part the balkanized nature of taxonomic science, as most systematists specialize on groups of related organisms and have limited scientific interactions with those studying other groups. Last year Jesse Ausubel and Paul Waggoner outlined some of the concerns raised by the growth of DNA barcoding in Barcoding Worries and Limits. The final item asks whether “it is too soon to standardize on a very few localities”. The salamander paper discussed in last week’s post seems to support this concern, as it implies that the relative rates of evolution of mitochondrial genes differ between animal groups. According to the paper, in salamanders COI evolves much more rapidly than other mitochondrial protein-coding genes.
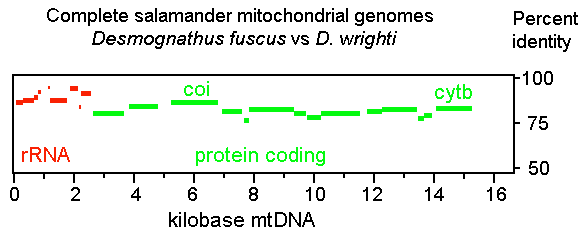
However, as detailed below, my analysis indicates that salamanders are unexceptional in terms of COI differences. I find evolutionary divergences are widely distributed in mitochondrial protein-coding genes and the patterning of these differences is similar in diverse invertebrate and vertebrate organisms, including salamanders. First, I used PipMaker (percent identity plot) to examine the distribution of sequence differences in complete mitochondrial genomes. A representative PipMaker plot, shown above, compares the mitochondrial genomes of 2 plethodontid salamanders, Desmognathus fuscus and D. wrighti, revealing a typical pattern of widely distributed sequence differences. As in most vertebrates, in salamanders COI is slightly LESS divergent (14%) than other protein-coding regions, including cytochrome b (CYTB) (17%).
Second, I compiled differences between closely-related species pairs in CYTB and COI. For historical reasons, CYTB has been the single most common locus used to analyze differences in vertebrate species, and COI has been the single most common single locus for invertebrates. As shown below, these genes show highly correlated differences in deeply divergent lineages of invertebrates and vertebrates, including salamanders.
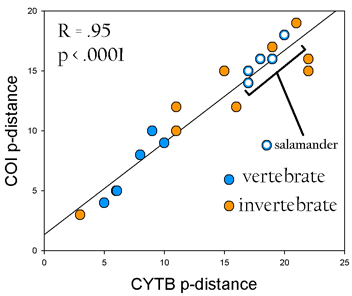
Species examined include Demospongiae (sponges); Platyhelminthes (tapeworms); Echinodermata (starfish); Arthropoda (ticks; fruit flies; mosquitos; shrimp); Mollusca (octopi; mantis shrimp) Vertebrata (turtles; eels; coelecanths; elephants; wolves; apes; and 5 pairs of congeneric salamanders)
These findings support standardizing on COI as the primary DNA barcode for multicellular animals. As discussed in earlier posts, there will of course be cases in which the primary barcode does not resolve closely-related species and a secondary barcode(s) may be needed.
DNA barcoding identifies invasive pests, appears ready for agricultural application
Pest and invasive species in agricultural crops and shipped goods pose enormous economic and biosecurity threats. Rapidly and reliably identifying pests and invasives in all life stages is likely to be one of the most economically important applications of DNA barcoding. In March 2006 Ann Entomol Soc Am 99: 204 Scheffer, Lewis, and Joshi from the USDA and the Philippine Department of Agriculture apply DNA barcoding to invasive leafminer flies in the Philippines.  They analyzed 258 specimens from the Philippines and compared these to a database of 307 sequences previously collected from worldwide populations. Bootstrap values for all species were 100%. In addition, a total of 7 distinct clusters
They analyzed 258 specimens from the Philippines and compared these to a database of 307 sequences previously collected from worldwide populations. Bootstrap values for all species were 100%. In addition, a total of 7 distinct clusters
with 98-100% bootstrap support were found in 3 “morphospecies”, suggesting discovery of new species, similar to findings of unrecognized biodiversity in all DNA barcode surveys to date. They conclude “DNA barcoding of economically and medically important species offers a powerful means of rapid identification”.
With DNA barcoding, all can identify this Liriomyza sativae pupae
COI evolves rapidly in salamanders, helping distinguish and discover species, still some taxonomists worry
Results so far in animals suggest the major limitation to DNA barcoding is very young species pairs. Young species pairs may not have accumulated enough differences in the 648 bp barcode region of COI to form distinct lineages in neighbor-joining analysis. Because overall rates of mitochondrial DNA evolution differ among animal groups, and the relative rates of specific mitochondrial genes also differ, it may turn out that COI barcoding will be more effective in separating young species pairs in some groups than in others.
20 My of genetic divergence and morphologic stasis
In April 2006 Systematic Biology Mueller compares complete mitochondrial genomes of 27 salamander species. The genetics of salamanders are particularly important as they are notoriously difficult to distinguish and classify due to morphologic stasis and frequent homoplasy. Many new species of salamanders have been discovered and described on the basis of mitochondrial divergence. Mueller found that COI has the highest rate of evolution among all mitochondrial genes in salamanders, suggesting it will be especially effective at resolving young species in this group. Despite these very encouraging results, the paper ends with a worried discussion about using COI for DNA barcoding of salamanders, because its evolutionary rate varies between lineages.
A better appreciation of the power of a standardized genetic approach for amphibians is Ron et al.’s paper in May 2006 Molecular Phylogenetics and Evolution which uses mitochondrial DNA to look at phylogeny and new species in the Neotropical tungara frog genus Engystomops. The authors observe that “the use of genetic markers in systematics has an enormous potential to facilitate the global inventory of biodiversity”, and conclude, based on their results and those in similar studies, that “the increasing use of molecular techniques will lead to the discovery of a vast number of species of Neotropical amphibians.”
New species descriptions could benefit from DNA barcodes
 A year ago in Science, Jones et al. described a new species of African monkey, Lophocebus kipunji, documenting their report with field observations, sound recordings, and photographs. This led to legalistic wrangling over whether the species could be said to exist if there was no specimen! Fortunately, the suspense is relieved by Davenport et al. in this month’s Science, in which they provide morphologic and DNA information based on a specimen recovered from a farmer’s trap. Although the authors relied on mitochondrial DNA evidence to establish the monkey belongs in a new genus, Rungwecebus, the actual sequence data is not listed among diagnostic characters in the species or genus description. Routine inclusion of DNA barcode sequences could improve the usefulness of formal species descriptions, assisting primate conservation efforts that monitor bushmeat trade, for example.
A year ago in Science, Jones et al. described a new species of African monkey, Lophocebus kipunji, documenting their report with field observations, sound recordings, and photographs. This led to legalistic wrangling over whether the species could be said to exist if there was no specimen! Fortunately, the suspense is relieved by Davenport et al. in this month’s Science, in which they provide morphologic and DNA information based on a specimen recovered from a farmer’s trap. Although the authors relied on mitochondrial DNA evidence to establish the monkey belongs in a new genus, Rungwecebus, the actual sequence data is not listed among diagnostic characters in the species or genus description. Routine inclusion of DNA barcode sequences could improve the usefulness of formal species descriptions, assisting primate conservation efforts that monitor bushmeat trade, for example.
Where is my barcode?
To rapidly map biodiversity, stomatopods suggest start with DNA
In “Estimating diversity of Indo-Pacific coral reef stomatopods through DNA barcoding of stomatopod larvae” (FirstCite early online publication in Proceedings Royal Society Biology) Paul Barber and Sarah Boyce, Boston University, look at  a what is thought to be a well-understood group, stomatopods, commonly known as mantis shrimp. Stomatopods are marine crustaceans distinct from true shrimp and are thought to include about 400 species worldwide. Like many marine species, they have a bipartite life cycle where dispersal is achieved through a planktonic larval developmental stage. However, larval stages are notoriously difficult to identify morphologically. Barber and Boyce first established a database of COI DNA barcodes from adult forms of nearly all known species. They then applied DNA barcoding to planktonic larvae collected in light traps at locations in the Pacific Ocean and Red Sea. Comparison of
a what is thought to be a well-understood group, stomatopods, commonly known as mantis shrimp. Stomatopods are marine crustaceans distinct from true shrimp and are thought to include about 400 species worldwide. Like many marine species, they have a bipartite life cycle where dispersal is achieved through a planktonic larval developmental stage. However, larval stages are notoriously difficult to identify morphologically. Barber and Boyce first established a database of COI DNA barcodes from adult forms of nearly all known species. They then applied DNA barcoding to planktonic larvae collected in light traps at locations in the Pacific Ocean and Red Sea. Comparison of  sequences from 189 larval forms revealed 22 distinct larval operational taxonomic units (OTUs), but a minority of these matched known species, suggesting that stomatopod species diversity is underestimated by a minimum of 50% to more than 150%. Their results support general use of DNA barcoding as a rapid, relatively-inexpensive first step in cataloging marine species with planktonic larvae. A similar approach has been applied on land by Smith et al “DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar“.
sequences from 189 larval forms revealed 22 distinct larval operational taxonomic units (OTUs), but a minority of these matched known species, suggesting that stomatopod species diversity is underestimated by a minimum of 50% to more than 150%. Their results support general use of DNA barcoding as a rapid, relatively-inexpensive first step in cataloging marine species with planktonic larvae. A similar approach has been applied on land by Smith et al “DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar“.
DNA barcode OTUs, such as found in these studies, are not equivalent to species descriptions and are not sufficient to establish systematic phylogeny. In my view, these studies indicate that DNA barcodes can be permanent indexers for filing and retrieving biologic information in the encyclopedia of life. Routinely incorporating DNA barcoding into biological surveys will enhance the long-term value of expensive field work.
JIE Celebrated 10th Anniversary
On Thursday, May 11, 2006 the Journal of Industrial Ecology celebrated its 10th anniversary and inclusion in the Science Citation Index with a seminar and festive program. Jesse was one of three honorees, for PHE’s work on Dematerialization, carried out with Robert Herman and Iddo Wernick.
Zooplankton sequenced at sea, illuminating life in the dark
 The deep oceans are the largest biotic space on earth, but remain largely unexplored. Census of Marine Life scientists recently trawled the Atlantic between the southeast US coast and the Mid-Atlantic ridge, focusing on the zone of perpetual darkness that lies below about 1000 m, to inventory and photograph the variety and abundance of zooplankton–the tiny sea animals that form a vital link in ocean food chains. The 20 day cruise completed April 20 is part of the ambitious gobal inventory of zooplankton by 2010 (Census of Marine Zooplankton (CMarZ), a Census of Marine life initiative. As reported in Science, for the first time DNA sequencing was performed on the rolling seas, telescoping into just three weeks what would normally represent years of laboratory work.
The deep oceans are the largest biotic space on earth, but remain largely unexplored. Census of Marine Life scientists recently trawled the Atlantic between the southeast US coast and the Mid-Atlantic ridge, focusing on the zone of perpetual darkness that lies below about 1000 m, to inventory and photograph the variety and abundance of zooplankton–the tiny sea animals that form a vital link in ocean food chains. The 20 day cruise completed April 20 is part of the ambitious gobal inventory of zooplankton by 2010 (Census of Marine Zooplankton (CMarZ), a Census of Marine life initiative. As reported in Science, for the first time DNA sequencing was performed on the rolling seas, telescoping into just three weeks what would normally represent years of laboratory work.
 According to Ann Bucklin, lead scientist for CMarZ and Head of the University of Connecticut Marine Sciences Department, “we are just starting to realize how little we know about species variety. We used to think we knew many species well, but the advent of DNA barcoding has radically altered that perception. Genetically distinctive species of zooplankton are being found with increasing frequency.”
According to Ann Bucklin, lead scientist for CMarZ and Head of the University of Connecticut Marine Sciences Department, “we are just starting to realize how little we know about species variety. We used to think we knew many species well, but the advent of DNA barcoding has radically altered that perception. Genetically distinctive species of zooplankton are being found with increasing frequency.”
Serious Games
Our longstanding interest in Serious Games extended during the past year to the Reinventing Public Diplomacy Through Games Competition organized by Josh Fouts and Doug Thomas at USC sponsored by the Lounsbery Foundation. First place went to PeaceMaker, a cross-cultural political video game simulation of the Israeli-Palestinian conflict which can be used to promote a peaceful resolution among Israelis, Palestinians and young adults worldwide. Congratulations to Asi Burak and PeaceMaker team members at Carnegie-Mellon.
A little history of our involvement with gaming: Swedish economist Ingolf Stahl first involved Jesse in interactive gaming in 1980-1981, when Ingolf, Jesse, Jennifer Robinson, and John Lathorp built a board game and two computer games about carbon dioxide emissions and global warming (described in Jennifer Robinson and Jesse H. Ausubel, A Game Framework for Scenario Generation for the CO2 Issue, Simulation and Games 14(3):317-344). The 1981 paper, Ingolf Stahl and Jesse H. Ausubel, Estimating the Future Input of Fossil Fuel CO2 into the Atmosphere by Simulation Gaming (IIASA Working Paper-81-107, published in Beyond the Energy Crisis-Opportunity and Challenge, Rocco.A. Fazzolare and Craig B. Smith, eds., Oxford, Pergamon, 1981) was the first paper to introduce the idea of greenhouse gas emission trading. Jesse’s 1988 book with Robert Herman, Cities and their Vital Systems: Infrastructure Past, Present, and Future, found its way to the brilliant young Will Wright, and Will made terrific use of it in his first 1989 release of SimCity, for which Jesse had the great opportunity to be a beta tester. Jesse wonders whether he still somewhere has the diskettes of the beta version. Will opened Jesse’s eyes to modern simulation and helpfully advised Jesse and Bill Massey on the creation of the university simulator, Virtual U . (For the game history: https://phe.rockefeller.edu/jesse/vupenn.pdf). Virtual U provided the kernel around which the enterprising Ben Sawyer (https://www.dmill.com) and Dave Rejeski nucleated the Serious Games initiative. Jesse still hopes to find a programmer who would take the 1981 Greenhouse Effect board game and turn it into an on-line game.