In October 2006 Conservation Biology, Rob DeSalle, American Museum of Natural History, comments on April 2006 CB piece on barcoding by Daniel Rubinoff, University of Hawaii (also see earlier Barcode Blog post on this article), and DeSalle’s commentary is followed by reply from Rubinoff. Such fun! DeSalle divides his analysis of DNA barcoding into its utility in “species identification” ie assigning specimens to known species, and “species discovery” ie formal descriptions of new species previously unknown to science. Both researchers cede the field of “species identification” to DNA barcoding (DeSalle: “a species identification system based on DNA sequences [would] be reliable, consistent, and rapid”; Rubinoff: “barcodes could be invaluable for speed and accuracy”).
Establishing reference libraries will be a major scientific achievement on the scale of the Human Genome Project. As these become available, the limiting factors for DNA barcoding will be cost and availability of sequence analysis, but as DeSalle observes, “it is not unreasonable to assume that DNA technology will advance to the stage where field-based diagnostics can be accomplished”.
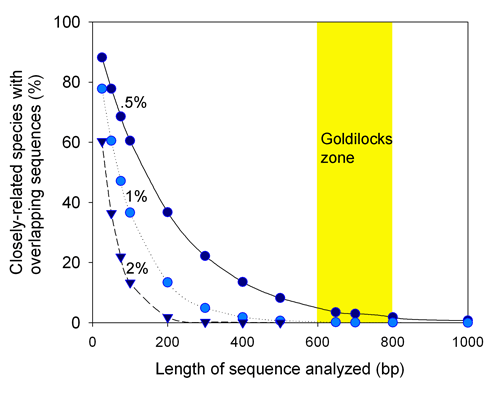
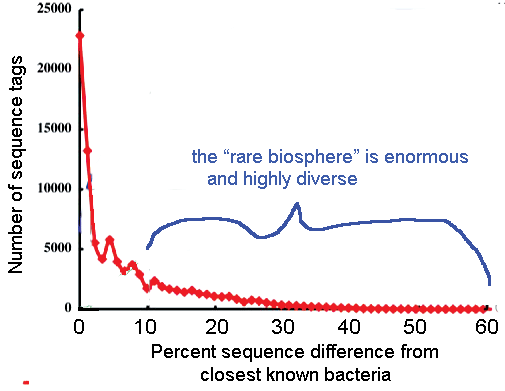
Since there are about 1.1 million named multicellular animal species, “species identification” is a vast area for scientific research and practical application of DNA barcoding. It seems likely that the 1.1 million known species includes most of the more abundant and wide-ranging species, and most that are of direct economic or scientific importance to humans. It is generally believed there are many more undescribed species than what has already been named. These may be largely rarer species with limited distributions (see earlier post on “rare microbial biosphere“). Population sizes and ranges in the undescribed biosphere, together with measures of genetic diversity (see last week’s post) might be interesting research areas.
The rest of the exchange centers on the role of DNA barcoding in “species discovery” ie formal descriptions of species previously unknown to science. Here both turn cautious, reserving an essential role for expert judgement. (DeSalle: “DNA sequence information in the absence of other corroborating evidence can never be used by itself as an indicator of species delimitation”. Rubinoff: “my opposition is to the practice by which species are known and identifiable only through a DNA barcode”.)
It may be that it takes a taxonomist to recognize a new species and that it is essential to use an integrated approach combining morphology, ecology, together with DNA sequence data. However, I am struck that in practice taxonomists often apply a “DNA-first” approach and that an untrained person could recognize most of the sequence clusters that correspond to species. A Google search with “new species” and “dna” turns up dozens of reports in which DNA sequence differences are the first and strongest evidence for cryptic species, including the 2 new shark species shown below.


It is surprising that the published description of the new shark species shown above did not include any DNA sequence data!
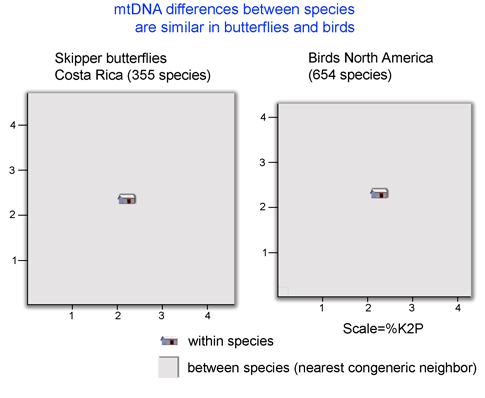
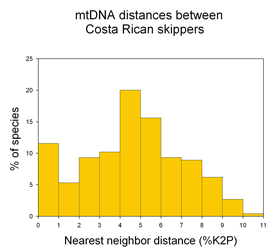
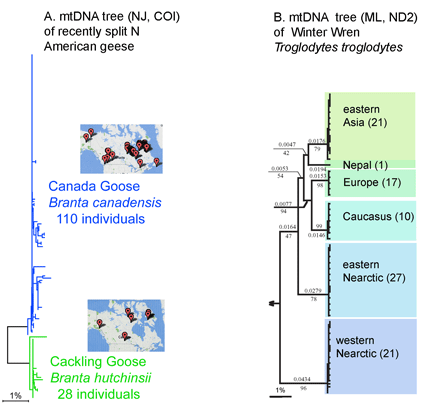
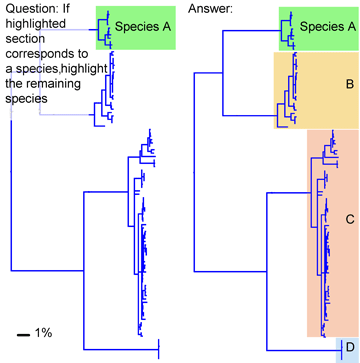
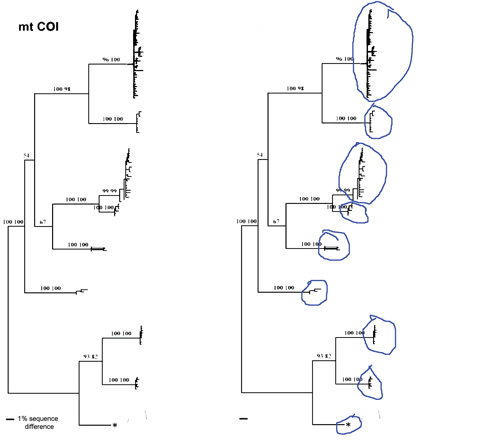
I close with a question about distance vs character comparisons of sequence data. Both authors assume that “distances” cannot be part of species descriptions, “characters” are needed. I wonder if this is a scientific fact, or one that reflects the social norms of taxonomy. As discussed in recent posts, neighbor-joining distance comparisons show most animal species as tight clusters, distinct from those of other species. Once a reference library is established, why not use distance clustering as a diagnostic, eg “species X COI barcodes lie within cluster 1439″?