News
New Yates, Stoeckle & Co paper relating eDNA production to surface area of organisms
The journal Environmental DNA publishes open access “Interspecific allometric scaling in eDNA production among northwestern Atlantic bony fishes reflects physiological allometric scaling” by Matthew C. Yates, Taylor M. Wilcox, M. Y. Stoeckle, and Daniel D. Heath. pdf here.
The paper finds that integrating allometry significantly improved correlations between organism abundance and metabarcoding read count relative to traditional metrics of abundance (density and biomass) for bony fishes. Future studies investigating the relationship between eDNA signal strength and metrics of fish abundance could improve by accounting for allometry; to this end, the paper develops an online tool that can facilitate the integration of allometry in eDNA/abundance relationships.
Interspecific allometry eDNA – an online tool
Explore how accounting for allometric scaling in environmental DNA (eDNA) shedding rates influences the correlation with organism count and abundance data. Users can upload their own datasets and use the slider to vary the allometric scaling coefficient (b). Allometry can be applied intra-specifically (among populations) or interspecifically (among species). Size-distribution data supplied to the function can also either be individual-level (e.g. a size distribution of individuals for each population) or population-level (e.g. the mean mass of individuals within a population).
Jesse podcasts on “Peak Human?”
Journalist/author Robert Bryce interviews Jesse Ausubel about PHE’s work on “peak human” and “peak humans.” The interview covers four dimensions of human performance: the physical (how far and fast can we go?), lifetime (how long can we live and how well?), cognitive (measures of intelligence and learning), and immunity (is our resistance to disease waning?). The podcast was recorded on December 7, 2022. For the audio and transcript, see the Bryce website, and also on YouTube.
Our paper on eDNA as bioassay of Anthropocene published
The new journal based in China, The Innovation, has published the Thaler-Ausubel-Stoeckle paper on Human and domesticated animal environmental DNA as bioassays of the Anthropocene in their “Out of the Box” category, where we like to be. We also post the pdf.
We thank Song Sun and Ke Chen for editorial assistance.
Summary: Human and domesticated animal sequences, commonly detected in environmental DNA (eDNA) metabarcoding studies, are routinely excluded from analysis. Here we suggest that reporting human and domesticated animal eDNA results might open new lines of investigation. For example, the relative abundance of human and domesticated animal eDNA as compared to that of wild vertebrate species might provide an index of human impact on local biota. Such an index could be applied to sites ranging from urban harbors to remote villages, and possibly to analyze historical samples. Various potential sources of contamination complicate the picture, but it should be possible to develop procedures that minimize risk of DNA introduction during collection and processing. Our near-term recommendation is to encourage inclusion of human and domesticated animal data in eDNA publications as an incentive for discovery, to lift quality controls, and to collectively contribute to new vistas that eDNA science might open.
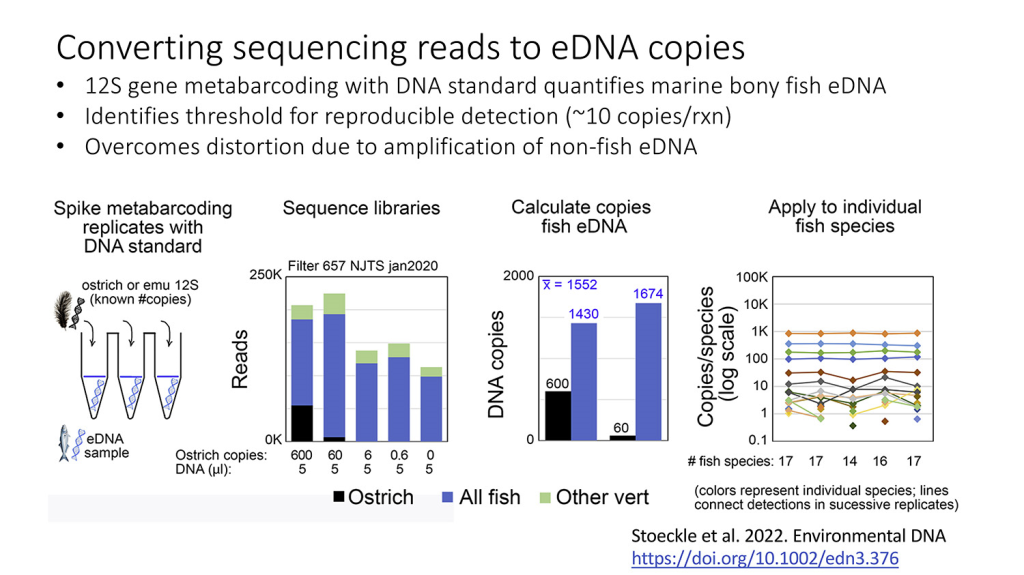
Our article on incorporating a known amount of non-fish DNA to allow better quantification of the fish DNA present in a seawater sample appears in the journal Environmental DNA.
Open Access
12S gene metabarcoding with DNA standard quantifies marine bony fish environmental DNA, identifies threshold for reproducible detection, and overcomes distortion due to amplification of non-fish DNA. Mark Y. Stoeckle, Jesse H. Ausubel, Michael Coogan, first published: 08 December 2022, https://doi.org/10.1002/edn3.376
While our paper focuses on fish, we believe the approach of “spiking” samples collected in nature with known amounts of DNA from a species that would not be present in the sample (such as ostrich) offers great promise for increasing the value of a wide range of aquatic DNA studies. The exhibit below shares some of the main points from the paper.
2007 Interview with Jesse cited in WSJ
A 5 Nov 2022 Wall Street Journal article by Holman Jenkins cites
JH Ausubel. Renewable and nuclear heresies . Int J Nucl Governance, Econ Ecol 1 (3): 229–243, 2007 and this interview with Robert Bryce also from 2007.
Thaler memo on “Distinguishing contamination from authentic human eDNA”
PHE guest investigator David Thaler summarizes “Ways in which contamination might be distinguished from authentic human eDNA” in a useful draft memo. Meeting this challenge matters greatly for using human eDNA as an assay of the Anthropocene, a subject of a forthcoming paper by Thaler, Ausubel, and Stoeckle.
Jesse’s Search for Leonardo’s Genome published
The journal Human Evolution has published Jesse Ausubel’s “The Search for Leonardo’s Genome,” an expanded, fully referenced version of a talk Jesse gave in June 2022 to a meeting of the American Academy of Arts & Sciences. The Academy Bulletin will publish the colloquial version in its winter issue.
Jesse H. Ausubel, The Search for Leonardo’s Genome, Human Evolution 37 3-4: 221-228, 2022; DOI: 10.14673/HE2022341106
Jesse’s Search for Leonardo’s Genome published in Human Evolution
The journal Human Evolution has published “The Search for Leonardo’s Genome” (Vol. 37 – n. 3-4 pp 221-228, 2022; DOI: 10.14673/HE2022341106), a fully referenced expansion of a talk Jesse Ausubel gave in June 2022 to a meeting of the American Academy of Arts and Sciences. The Academy’s Bulletin will published the colloquial version in its winter issue.
New code for analyzing eDNA sequences using DADA2 pipeline
For the past year MIchael Coogan, now a grad student in marine science at the U. of New Hampshire, has helped Mark Stoeckle and PHE with improved software for our eDNA studies. See summary below. A pdf of the R code is available here. If you have questions, please write to Mark.
The goal is to adapt the DADA2 pipeline to Mark Stoeckle’s 12S experiment. Sample sequences will be identified using 12S reference file containing sequences of 262 unique vertebrates found around New York. The starting point is a set of Illumina-sequenced paired-end fastq files that have been split (or demultiplexed) by sample and which have barcodes/adapters already removed. The end product will be a sequence table, analogous to the ubiquitous “OTU table”, which records the number of times sample sequences were observed in each sample. The key difference between the output of DADA2 and standard OTU analyses is that DADA2 infers sample sequences exactly rather than clustering sequences into fuzzy OTUs which hide and complicate biological variation.