As Dirk Steinke’s recent blog post demonstrated, since the seminal 2003 Proc Royal Soc London B Biol Sci paper by Hebert, Cywinska, Ball, and DeWaard, barcoders around the world have been generating scientific papers at a steadily growing pace. For more on the big picture, here I share three barcode stat visuals put together in preparation for the Third European Congress for the Barcode of Life (ECBOL3) in September.
Q: How many specimens have been barcoded?
A: A lot.
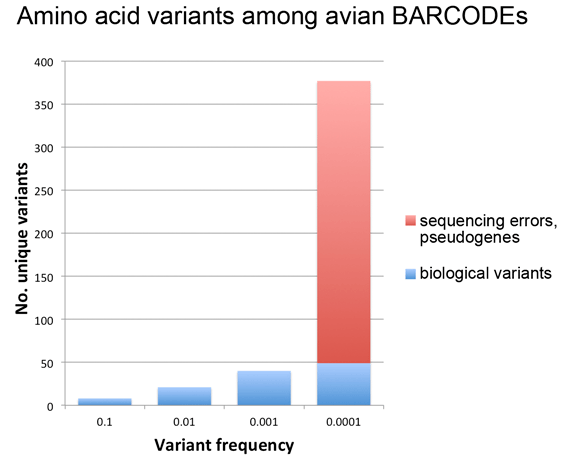
As of September 2012, about 600,000 specimens have barcode records in GenBank, about half of which qualify for BARCODE[keyword] based on CBOL data standards. This reminds me to recognize the special challenges barcoding has as a genomics project–the target number of specimens is enormous and each requires expert identification and long-term storage in a museum or herbarium.
In addition to GenBank/BOLD public records, at the time of the survey there were another 1.2 million barcode records in BOLD which lack species names. Probably most represent what Rod Page called “dark taxa“–difficult to identify specimens from undescribed species. It is an unsolved puzzle how much effort to devote to barcoding specimens that can’t or haven’t been identified to species. On the one hand this approach speeds species discovery, as documented in blog post cited above; on the other hand, many specimens will wait a very long time for the right taxonomist to come along and in the meantime the sequences alone may not be very useful to science. I should point out that for many dark barcodes, the sequences are public, are labeled with an order level identifier (e.g. Vertebrata) and BIN (see below), and include specimen photographs.
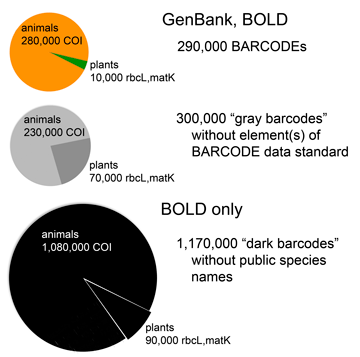 One possible solution is assigning “names” based on barcode sequences themselves, such as Barcode Index Number (BIN) system instituted in BOLD. This sidesteps the wait for an expert human to assign a traditional Latin binomial but does not link the sequence to other biological information about the organism the way a species name does. Researchers recently estimated there are about 8.7 million eukaryotic species, of which about 2 million are named (Mora et al PLoS Biol 2011). Given the very large array of undescribed (mostly small) life, how should barcoders proceed? The Human Genome Project seized on what was a radical idea and technologically difficult at the time–namely, sequencing the whole genome rather than just the expressed genes. Does an analogous approach of sequencing the whole eukaryotic biome of 8.7 million predicted species make sense? Let’s say we had sequences for all these forms–what new knowledge or capabilities would we have? I favor a stepwise approach focused on barcoding organisms already named, particularly those are already in collections and those important to society. There will be plenty of species discovery along the way.
One possible solution is assigning “names” based on barcode sequences themselves, such as Barcode Index Number (BIN) system instituted in BOLD. This sidesteps the wait for an expert human to assign a traditional Latin binomial but does not link the sequence to other biological information about the organism the way a species name does. Researchers recently estimated there are about 8.7 million eukaryotic species, of which about 2 million are named (Mora et al PLoS Biol 2011). Given the very large array of undescribed (mostly small) life, how should barcoders proceed? The Human Genome Project seized on what was a radical idea and technologically difficult at the time–namely, sequencing the whole genome rather than just the expressed genes. Does an analogous approach of sequencing the whole eukaryotic biome of 8.7 million predicted species make sense? Let’s say we had sequences for all these forms–what new knowledge or capabilities would we have? I favor a stepwise approach focused on barcoding organisms already named, particularly those are already in collections and those important to society. There will be plenty of species discovery along the way.
Other dark barcodes are simply records for which the researchers have assigned a species name but are not posting it publicly. The importance of making sequence data public quickly was recognized at the 4th International Barcode of Life Conference held in Aidelaide last year (for one example of rapid publication of DNA barcode data see Schindel 2011 ZooKeys). Open access, data sharing, and transparency have been embraced by many scientific fields and their funders and I hope barcoders already have or are moving to adopt these principles.
Q: How many species have been barcoded?
A: A lot.
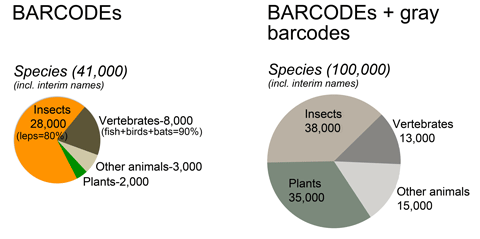
GenBank holds barcode sequences for about 100,000 species, mostly insects, vertebrates, and plants, and about 40,000 qualify for BARCODE keyword. Nearly all BARCODE records so far are from animals, mostly lepidoptera and vertebrates.
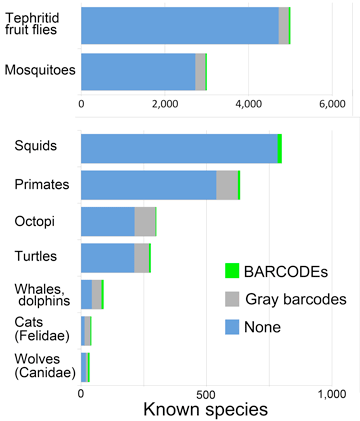 Q: What groups important to science or society have few barcodes?
Q: What groups important to science or society have few barcodes?
A: Quite a few.
These suggest opportunities for scientific progress and grant support. They include human and animal disease vectors, agricultural pests, threatened and endangered species, and notable marine groups.
Powerpoint of slides available here.