 Robert Koch (1842-1910), father of medical microbiology, isolated agents of mankind’s major plagues: Vibrio cholera, Bacillus anthracis (anthrax and bubonic plague), and Mycobacterium tuberculosis. He laid down four conditions, “Koch’s postulates“, for establishing that an organism is the agent of disease, and subsequent generations of researchers applied these principles to determine the etiology of a multitude of infectious diseases. One legacy of Koch’s postulates was that isolation of organisms in pure culture became the backbone of diagnostic and research microbiology.
Robert Koch (1842-1910), father of medical microbiology, isolated agents of mankind’s major plagues: Vibrio cholera, Bacillus anthracis (anthrax and bubonic plague), and Mycobacterium tuberculosis. He laid down four conditions, “Koch’s postulates“, for establishing that an organism is the agent of disease, and subsequent generations of researchers applied these principles to determine the etiology of a multitude of infectious diseases. One legacy of Koch’s postulates was that isolation of organisms in pure culture became the backbone of diagnostic and research microbiology.
A century later, genetics has replaced culture as the essential framework for exploring microbial life. Metagenomic analysis of environmental samples, including from anatomic sites, has identified an unsuspected plethora of organisms, most of which are unculturable, at least under standard laboratory conditions. Even for organisms that can be grown in the laboratory, genetic detection is often the preferred diagnostic method, including for example detection of HIV, Neisseria gonorrhea, and Chlamydia sp. Following Carl Woese’s early lead (PNAS 1977, 74:5088), microbiologists have generally included a standard locus, 16s rRNA, in genetic work, enabling phylogenetic trees spanning the diversity of life, and allowing each new isolate to be analyzed in conjunction with the work of others (as of 24 oct 2008, 75,257 16S rRNA sequences in GenBank).
Are genetic methods equally necessary for eukaryotes? In October 2008 Mol Ecol researchers from Cardiff University analyze mitochondrial COI differences among nine species of British lumbricid earthworms which were first described between 1758 and 1843, over 150 years ago. Partial COI sequences (a 582 bp segment which overlaps 648 bp DNA barcode region) from 71 individuals showed 2-5 deeply divergent clusters (average 13-15% sequence difference) in 4 of the 8 multiply-sampled species, and small divergences within each cluster, “indicative of the presence of multiple previously undescribed species”. COI sequences from 270 individuals of one species, Allolobophora chlorotica, collected at 24 British and 5 mainland European sites showed 5 divergent clusters and surprisingly no clear geographic distribution pattern; over half the sites had 2 or more lineages, and one site had 4 lineages. As expected the same clusters were found by comparing another mitochondrial gene, 16s rRNA. Two of the lineages were found only in green color morphs; prior work indicated this form has distinct ecological preferences compared to pink morph Allo. chlorotica and that F1 hybrids are sterile, suggesting species status. As an aside, if earthworm specialists find morphological and ecological differences and mating incompatibility, why not designate as distinct species? As another example, two forms of European corn borer Ostrinia nubilalis are sympatric, genetically distinct, develop on different host plants, have different mating pheromones, and exhibit >95% reproductive isolation, yet are described as “host races” rather than separate species (Science 2005, 308:258). It sometimes seems there is an arbitrary aspect of how species status is awarded, or perhaps the process is slow.
To see if mtDNA clusters were also reflected in nuclear genome, King et al performed AFLP (amplified fragment length polymorphism) mapping on 4-12 individuals from each of the 5 lineages. The nuclear results corresponded exactly to COI clusters except that the 2 green morph forms could not be distinguished, suggesting these are either a single interbreeding species (despite 14% mtCOI sequence difference!) or are young species which have not yet accumulated differences in nuclear DNA. It is hard to see how a 14% sequence difference could accumulate in mtDNA without accompanying nuclear changes, so I wonder if one of the genetic forms might reflect a relatively recent introgression from another earthworm species which has not yet been sequenced. It will be interesting to see whether the two green morph lineages, which were often found together at the same site, show assortative mating or restricted fertility. The authors conclude “extraordinary species-level genetic diversity was revealed among the British earthworms”….”four of nine ecologically generalist earthworms are probably complexes of multiple cryptic species”. And finally “further earthworm research in areas such as ecology and ecotoxicology, should be conducted in the knowledge that there are multiple cryptic species within many earthworm species”.
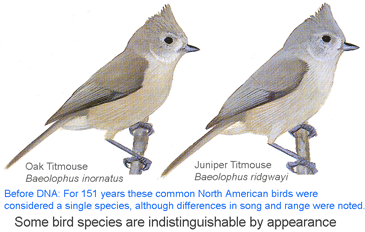 I conclude that genetics is equally essential for eukaryotic taxonomy as for microbiology. I believe there is no getting around the need to genetically reexamine most or all of the species named in the past 200 years to see if what we recognize as single and distinct species are really so. If there can be cryptic species in large visible animals such as birds, and males and females can be given different species names in fish, then there must be many more such oversights among the less easily observed. A standardized approach (ie DNA barcoding) is the most expeditious way forward and will leave a permanent marked trail that can easily be followed by non-experts who wish to identify their specimens. As in bacteria, standardizing on a single locus (ie barcode region COI for animals) enables new work to be seamlessly combined with old, leveraging its value (497,851 barcode records from 48,459 species in BOLD so far). Regarding higher-level evolutionary relationships, I find routine dismissal based on mathematical modeling of mtDNA single-locus trees, but not much effort to see what the potential is. Perhaps translated amino acid sequences and/or GC content can be informative for deeper branches, and nucleotide sequences for family- and generic-level relationships. At the very least, mtDNA trees serve to generate hypotheses, which can be corroborated or disproved by more extensive genetic, morphologic, ecologic, behavioral, or fossil record data.
I conclude that genetics is equally essential for eukaryotic taxonomy as for microbiology. I believe there is no getting around the need to genetically reexamine most or all of the species named in the past 200 years to see if what we recognize as single and distinct species are really so. If there can be cryptic species in large visible animals such as birds, and males and females can be given different species names in fish, then there must be many more such oversights among the less easily observed. A standardized approach (ie DNA barcoding) is the most expeditious way forward and will leave a permanent marked trail that can easily be followed by non-experts who wish to identify their specimens. As in bacteria, standardizing on a single locus (ie barcode region COI for animals) enables new work to be seamlessly combined with old, leveraging its value (497,851 barcode records from 48,459 species in BOLD so far). Regarding higher-level evolutionary relationships, I find routine dismissal based on mathematical modeling of mtDNA single-locus trees, but not much effort to see what the potential is. Perhaps translated amino acid sequences and/or GC content can be informative for deeper branches, and nucleotide sequences for family- and generic-level relationships. At the very least, mtDNA trees serve to generate hypotheses, which can be corroborated or disproved by more extensive genetic, morphologic, ecologic, behavioral, or fossil record data.
 As of October 11, 2008 researchers have deposited 14,594 DNA barcodes in BOLD representing 2,586 avian species, 26% of world’s 9,933 birds. You can browse taxonomic coverage to date at
As of October 11, 2008 researchers have deposited 14,594 DNA barcodes in BOLD representing 2,586 avian species, 26% of world’s 9,933 birds. You can browse taxonomic coverage to date at 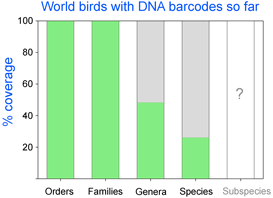 How far along are researchers toward mapping COI barcode resolution of avian species? Birds are of particular interest because species limits are generally well-defined, supported by a wealth of morphologic, ecological, behavioral, and other genetic data. Looked at regionally, there is good coverage in northern North America, parts of Central and South America, western Europe, Korea, and New Zealand, so it should be possible to see how well COI barcodes distinguish among local species in these areas. Published studies so far show >95% resolution of named species and have identified genetically divergent clusters which may represent unrecognized cryptic or “hidden” species (
How far along are researchers toward mapping COI barcode resolution of avian species? Birds are of particular interest because species limits are generally well-defined, supported by a wealth of morphologic, ecological, behavioral, and other genetic data. Looked at regionally, there is good coverage in northern North America, parts of Central and South America, western Europe, Korea, and New Zealand, so it should be possible to see how well COI barcodes distinguish among local species in these areas. Published studies so far show >95% resolution of named species and have identified genetically divergent clusters which may represent unrecognized cryptic or “hidden” species (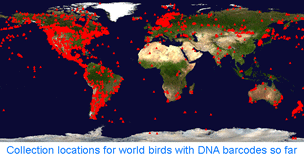 Looked at globally, there is 100% coverage of 104 polytypic genera (having 2 or more species) representing 324 birds, so this should include the sister species and/or “nearest neighbors” for these, plus there are 853 monotypic genera (having only 1 species) in world birds, which are likely or known to be genetically divergent from birds in other genera. In addition, there are likely many other sister species or “nearest neighbors” within the remaining 1,982 birds with DNA barcodes so far (for example, BOLD includes 28 of 29 Dendroica sp wood warblers). It would be interesting to look at the nearest neighbor differences within the global data set. To my knowledge, comparisons among regions with COI barcode data have been not been published. My impression based on other avian genetic work is that named taxa in different biogeographic regions are genetically distinct, plus there are many unrecognized genetic divisions within species that range across biogeographic regions. I look forward to trans-regional and global comparisons!
Looked at globally, there is 100% coverage of 104 polytypic genera (having 2 or more species) representing 324 birds, so this should include the sister species and/or “nearest neighbors” for these, plus there are 853 monotypic genera (having only 1 species) in world birds, which are likely or known to be genetically divergent from birds in other genera. In addition, there are likely many other sister species or “nearest neighbors” within the remaining 1,982 birds with DNA barcodes so far (for example, BOLD includes 28 of 29 Dendroica sp wood warblers). It would be interesting to look at the nearest neighbor differences within the global data set. To my knowledge, comparisons among regions with COI barcode data have been not been published. My impression based on other avian genetic work is that named taxa in different biogeographic regions are genetically distinct, plus there are many unrecognized genetic divisions within species that range across biogeographic regions. I look forward to trans-regional and global comparisons! In
In  In
In  In
In  An article in
An article in  In
In 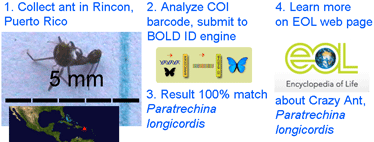 As a test of how DNA barcoding might work for the interested ant novice, I collected the tiny specimen at left in Rincon, Puerto Rico, and submitted its COI barcode to
As a test of how DNA barcoding might work for the interested ant novice, I collected the tiny specimen at left in Rincon, Puerto Rico, and submitted its COI barcode to