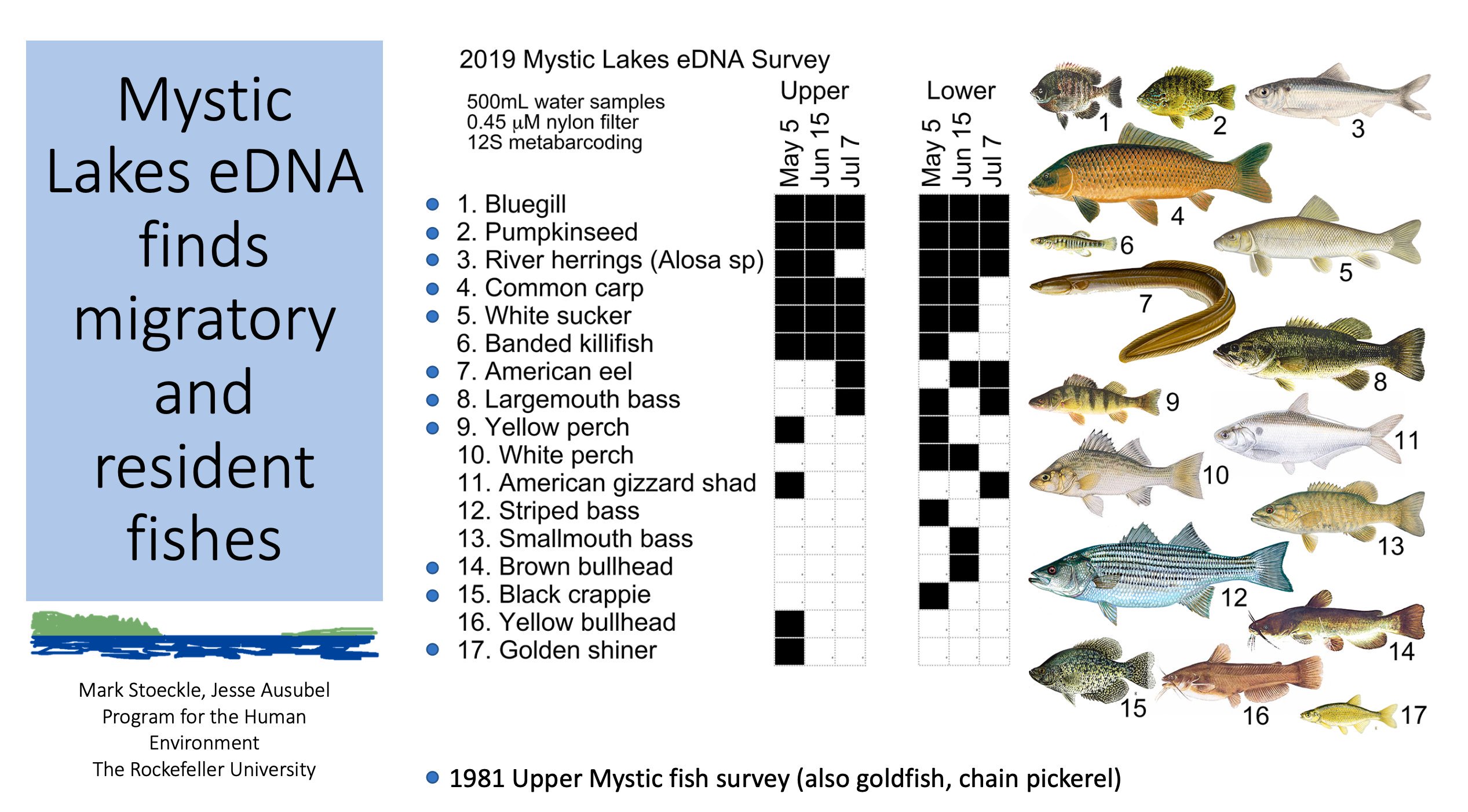
The Deep Carbon Observatory (DCO), which Jesse co-founded in 2009, is completing its initial decade. Jesse wrote a brief history of the program, including its origin in the work of the late Tommy Gold.
A press release summarizes some of the decadal achievements, celebrated at a large conference in Washington DC 24-26 October 2019, as does the Deep Carbon Observatory’s decadal report, a 50-page document released in October 2019.
Several books summarize the work of the DCO. Published by Cambridge University Press in October 2019, Deep Carbon: Past to Present, is a 684-page collection of 20 chapters by many authors edited by Beth N. Orcutt (Bigelow Lab, Maine, USA), Isabelle Daniel (University of Lyon, France), and Rajdeep Dasgupta (Rice University, Texas, USA). Aimed at sharing recent progress with the peer scientific community, the book is also available open access. The volume pairs with the 2013 volume Carbon in Earth, which aimed to provide the baseline of knowledge at the outset of the Deep Carbon Observatory program.
Published by W. W. Norton & Company in June 2019, Robert Hazen’s 288-page Symphony in C: Carbon and the Evolution of (Almost) Everything offers a popular tour of the work of the Deep Carbon Observatory including cameo portraits of many researchers involved in the program.
Finally, English historian of science Simon Mitton, has
completed the first history of deep carbon science, Carbon from Crust to Core: A
Chronicle of Deep Carbon Science.
Mitton’s 400-page book, to be published by Cambridge University Press in
early 2020, identifies key discoveries, impacts of new knowledge, and roles of
deep carbon scientists and their institutions from the 1400s to the present.