 DNA-based species descriptions could enable a catalog of life on Earth. Without some sort of automated approach, I believe this goal is unattainable. Insects are a good place to start testing an automated sequence-based approach, as there are about 1 million insect species already described, and probably several million more to go. In upcoming August 2006 Systematic Biology Pons et al examine genus Rivacindela tiger beetles in Australia, providing an explicit test of a DNA sequence-based approach to defining species. They analyzed 468 individuals from 65 sites, using sequence data from 3 mitochondrial genes including DNA barcode region of COI, and found sequence variation was strongly partitioned between 46 or 47 putative species, using a novel tree-based, quantitative method of species recognition based on fixed unique diagnostic characters. Most (40 to 43) of the species entities were recovered by analyzing the three gene regions separately; COI alone produced the closest match to the full data set. The putative species defined by sequence data exhibited biological properties of species in terms of geographic ranges and known morphologic characters. Average divergence within species was .5%, much lower than average among species of 6.3% and between sister species of 2.2%. The sequence analysis took 3 days on a desktop computer, so if this approach proves useful, it can be a benchmark for testing faster methods.
DNA-based species descriptions could enable a catalog of life on Earth. Without some sort of automated approach, I believe this goal is unattainable. Insects are a good place to start testing an automated sequence-based approach, as there are about 1 million insect species already described, and probably several million more to go. In upcoming August 2006 Systematic Biology Pons et al examine genus Rivacindela tiger beetles in Australia, providing an explicit test of a DNA sequence-based approach to defining species. They analyzed 468 individuals from 65 sites, using sequence data from 3 mitochondrial genes including DNA barcode region of COI, and found sequence variation was strongly partitioned between 46 or 47 putative species, using a novel tree-based, quantitative method of species recognition based on fixed unique diagnostic characters. Most (40 to 43) of the species entities were recovered by analyzing the three gene regions separately; COI alone produced the closest match to the full data set. The putative species defined by sequence data exhibited biological properties of species in terms of geographic ranges and known morphologic characters. Average divergence within species was .5%, much lower than average among species of 6.3% and between sister species of 2.2%. The sequence analysis took 3 days on a desktop computer, so if this approach proves useful, it can be a benchmark for testing faster methods.
News
Milan CoML
Milan’s excellent newspaper, Il Corriere della Sera, ran a colorful article about discoveries of the Census of Marine Life on 13 June 2006 featuring Jesse and Roberto Danovaro. Thanks to Fabio Caporizzi for interesting the Italian media.
Beginning to build a neotropical bird species index with DNA barcodes
 DNA barcodes index species. In most animal species studied so far, mtDNA differences within species are much smaller than those between species. As a result, species appear as distinct clusters in a simple neighbor-joining tree of COI barcodes. The uniformity of this patterning gives confidence that a DNA barcode library based on relatively few individuals per species will be a reliable index for assigning unknown specimens to known species. Although we are just at the beginning of compiling barcodes, and although we need phylogenetically-informed mathematical analysis about how to define clusters particularly in groups not well-studied, I am struck by how obvious most species clusters are. There are of course exceptions and limits (hybridization, young species, slow mitochondrial DNA evolution) but it is likely that someone with no knowledge other than a neighbor-joining tree of DNA barcodes could reconstruct most species categories, although they wouldn’t know anything about the biology of the organisms. This suggests viewing DNA barcoding as a diagnostic tool that links to biological knowledge, just as a laboratory test is used to detect HIV for example, and thereby point to a large body of biological knowledge.
DNA barcodes index species. In most animal species studied so far, mtDNA differences within species are much smaller than those between species. As a result, species appear as distinct clusters in a simple neighbor-joining tree of COI barcodes. The uniformity of this patterning gives confidence that a DNA barcode library based on relatively few individuals per species will be a reliable index for assigning unknown specimens to known species. Although we are just at the beginning of compiling barcodes, and although we need phylogenetically-informed mathematical analysis about how to define clusters particularly in groups not well-studied, I am struck by how obvious most species clusters are. There are of course exceptions and limits (hybridization, young species, slow mitochondrial DNA evolution) but it is likely that someone with no knowledge other than a neighbor-joining tree of DNA barcodes could reconstruct most species categories, although they wouldn’t know anything about the biology of the organisms. This suggests viewing DNA barcoding as a diagnostic tool that links to biological knowledge, just as a laboratory test is used to detect HIV for example, and thereby point to a large body of biological knowledge.
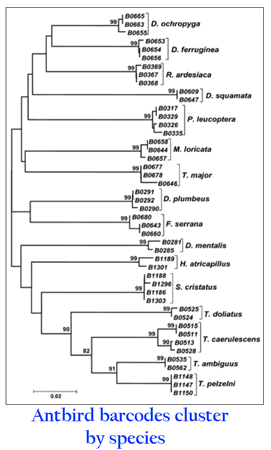
There are more bird species in the Neotropics than anywhere else. Over 4,000 of the approximately 10,000 world bird species live in South and Central America and the Caribbean, including over 3000 endemics. The large number of speciose families and the fact that intraspecific genetic variation is generally thought to be greater in the tropics than in temperate regions (eg Balakrishnan 2005 Syst Biol 54:689) might challenge DNA barcoding. In what I believe is the first explicit application of DNA barcoding to Neotropical birds (Vilaca 2006 Revista Brasileira Ornitologia 14:7) researchers analyzed 16 species of antbirds in the Atlantic Forest region of southeastern Brazil, with half of specimens obtained as blood samples from birds in the field. All species form distinct clusters in a neighbor-joining tree with 99% bootstrap support, including the recently split pair Thamnophilus pelzelni (shown above) and T. ambiguus. Maximum intraspecific variation is less than 1% except in T. caerulescens which shows 2 distinct lineages, highlighting a good candidate for further study.
Mitochondrial DNA analysis regularly reveals new species, supporting DNA to lead the way
There are so many examples of new animal species found through mitochondrial DNA analysis that I believe this should be a routine part of species descriptions. Using morphology alone, taxonomists have often overlooked species that are readily apparent on mitochondrial DNA analysis, including in what should be ideal circumstances using intact adult specimens of large, abundant, and/or economically important organisms. Morphologic characters have been found in some cases but only after DNA has led the way, indicating the discovery process would have been much faster if DNA were analyzed at the beginning. Speed is likely a good way to attract funding, as the public will want the fastest and therefore most economical approach. Mitochondrial DNA analysis can also help extinguish synonomies which have persisted in literature for decades (eg Siddall and Budinoff 2005. Conservation Genetics 6:467).
Under current practice, species recognition whether big or small can be slow (see also earlier post with timeline for discovery of New York Central Park centipede).
 Do you see a new species anywhere? Baleen whale specimen collected in 1976, new species description 27 years later based in part on mitochondrial DNA characters (Wada et al. 2003. Nature 426:278)
Do you see a new species anywhere? Baleen whale specimen collected in 1976, new species description 27 years later based in part on mitochondrial DNA characters (Wada et al. 2003. Nature 426:278)

Does this look like 1904? Honeybee mite Varroa jacobsoni described in 1904. In 1970’s, worldwide epidemic infestation of honeybees presumed due to V. jacobsoni began in Asia. 30 years later, epidemic discovered to be due to a new species, V. destructor (Anderson and Trueman 2000. Exp Appl Acarology 24:165). Species description based on mitochondrial DNA divergence; no morphologic characters other than body size.
Grapefruit-sized DNA sequencer in development
 With funding from the Gordon and Betty Moore Foundation, researchers at Reveo, Inc. and the University of Washington are collaborating on developing a grapefruit-sized sequencer. It uses electronic and photonic effects rather than liquid chemistry and could potentially sequence an entire genome for pennies.
With funding from the Gordon and Betty Moore Foundation, researchers at Reveo, Inc. and the University of Washington are collaborating on developing a grapefruit-sized sequencer. It uses electronic and photonic effects rather than liquid chemistry and could potentially sequence an entire genome for pennies.
In 2002, Godfray recognized that “in 10 or 20 years time it will be simpler to take an individual organism and get enough sequence data to assign it to a “sequence cluster” (equivalent to species) than to key it down using traditional methods” (Godfray 2002 Nature 417:17). That future is getting closer.
Here is your sequencer, sir
Brian Appleyard Article
The 11 June 2006 Londay Sunday Times magazine ran a feature article by Brian Appleyard on technological solutions to environmental problems that quotes Jesse several times.
Undiscovered species lurking in museum drawers
 Four whales stranded on the coast of California in the 1970s were identified as Hector’s beaked whale Mesoplodon hectori (Mead 1981 J Mammalogy 62:430). 20 years later mitochondrial DNA analysis revealed these 4 specimens, and a fifth stranded in 1997, to be a new species, designated Perrin’s beaked whale Mesoplodon perrini (Dalebout et al 2002 Marine Mammal Sci 18:577). The presence of a new species was first recognized by researchers compiling a database of mitochondrial DNA to assist in species identification. The formal species description includes diagnostic molecular characters, helping integrate DNA sequence data with classical taxonomy. As emphasized by Rob DeSalle and others, there is a need to interweave phylogeny and classical taxonomy, which can be met by including DNA sequences routinely used for evolutionary analysis as diagnostic characters in species descriptions (DeSalle et al. 2005 Phil Trans Royal Soc B 360:1905).
Four whales stranded on the coast of California in the 1970s were identified as Hector’s beaked whale Mesoplodon hectori (Mead 1981 J Mammalogy 62:430). 20 years later mitochondrial DNA analysis revealed these 4 specimens, and a fifth stranded in 1997, to be a new species, designated Perrin’s beaked whale Mesoplodon perrini (Dalebout et al 2002 Marine Mammal Sci 18:577). The presence of a new species was first recognized by researchers compiling a database of mitochondrial DNA to assist in species identification. The formal species description includes diagnostic molecular characters, helping integrate DNA sequence data with classical taxonomy. As emphasized by Rob DeSalle and others, there is a need to interweave phylogeny and classical taxonomy, which can be met by including DNA sequences routinely used for evolutionary analysis as diagnostic characters in species descriptions (DeSalle et al. 2005 Phil Trans Royal Soc B 360:1905).
I find it remarkable that a species as large as a whale can languish unrecognized in a collection. How many new species lurk in museum drawers awaiting discovery? A comprehensive DNA barcode survey of a tissue collection could be relatively inexpensive, as DNA isolation from tissues is generally simple and and sequencing costs are approaching a $1 a specimen. If whales can hide in museums for 20 years, there must be a multitude of new species already collected that will be uncovered by DNA analysis.
Learning how to apply DNA barcoding to species discovery
In 7 June 2006 Systematics and Biodiversity Andrew Brower examines application of DNA barcodes to identifying and defining new species. His Perspective piece “Problems with DNA barcodes for species delimitation: ‘ten species’ of Astraptes fulgerator reassessed (Lepidoptera: Hesperiidae)” reviews Hebert et al’s 2004 PNAS paper “Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. My short summary is that we are still learning about how best to use DNA barcodes for identifying new species. In an April 2006 post responding to another worry piece I wrote: “DNA barcoding is a taxonomic tool for a) assigning specimens to known species and b) speeding discovery of new species. More work is needed to determine the best use of DNA barcodes in species discovery.”
 Brower’s article argues about how many Astraptes species are supported by the sequence data, worries about possible consequences of the rise of DNA barcoding, and misses the scientific opportunity to examine this very young species complex with a large set of morphologic, ecologic, and sequence data. He concedes “there are probably at least three species” but declines to put forth what criteria can be used to delimit species. I was surprised to read that the question of whether two sequences “belong to the ‘same species’ is a metaphysical one”.
Brower’s article argues about how many Astraptes species are supported by the sequence data, worries about possible consequences of the rise of DNA barcoding, and misses the scientific opportunity to examine this very young species complex with a large set of morphologic, ecologic, and sequence data. He concedes “there are probably at least three species” but declines to put forth what criteria can be used to delimit species. I was surprised to read that the question of whether two sequences “belong to the ‘same species’ is a metaphysical one”.
A brief summary may be helpful. The PNAS Astraptes paper reports on 25 years of ecological and morphological work on Astraptes fulgerator larva and adults in a conservation area in northwestern Costa Rica. Approximately 40% of the 2,592 A. fulgerator caterpillars collected over this interval were successfully raised to adult; DNA barcodes were derived from the adult specimens. Differences in larval morphology and food plants and in some cases subtle differences in adult morphology suggested the presence of 6-7 cryptic species. When ADDED to this very large natural history knowledge base, DNA barcoding supported these inferences and highlighted another possible 3 or 4 species. The number of individuals sequenced is larger (4-103 per putative species; 7 of 10 with more than 40 specimens sequenced) than in any other single species study I am aware of. Despite the large sample sizes and detailed information on biological co-variation in ecological and morphological traits, Brower seems to read the paper as defining species based on DNA barcodes. According to his own analysis, NJ bootstrap recovered 7 species and cladistic haplotype analysis recovered 8, albeit with low statistical support. The argument over whether some of the putative species are established to be distinct biological entities is important for the Astraptes specialist, but misses the point of how well barcode lineages in the neighbor-joining tree match differences in biology in this VERY YOUNG species complex. To my reading, the findings suggest that if one were starting over, or starting with a unstudied group, using DNA barcoding as a first step would be the fastest way for taxonomists to sort specimens and find new species.
Newsprint
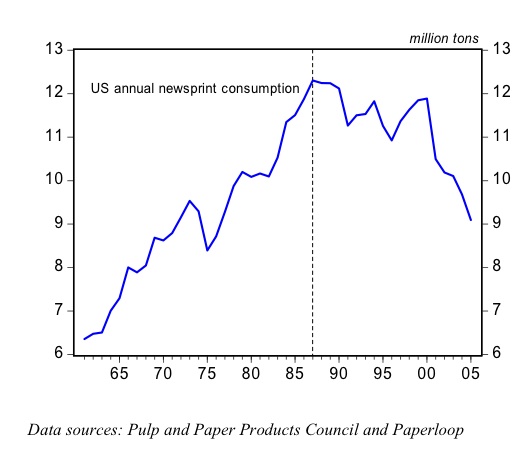
Finnish analyst Lauri Hetemaki documents a powerful example of Dematerialization in the U.S. newsprint market. For more information, see, Hetemäki, L. 2005, Chapter 6.2.2 “The U.S. newsprint market”, pp. 77-80, in ICT and Communication Paper Markets, Chapter 6 in Hetemäki, L. & Nilsson, S. (eds), . IUFRO World Series, Vol. 18, August 2005, Vienna.
Long Island CoML Talk
Friday 9 June 2006 Jesse presents a progress report on the Census of Marine Life to a Long Island chapter of The Nature Conservancy.