The biological universe is much larger and more diverse than we thought. In three papers in March 2007 PLoS Biology, scientists report on a genetic survey of microbial diversity in the world’s oceans. A large collaboration, the Global Oceanic Sampling (GOS), led by Craig Venter, analyzed microbial DNA collected by filtering seawater at 250 sites along a several thousand kilometer transect from the North Atlantic, through the Panama Canal, around the Galapagos Islands, ending in the Cocos Islands of the South Pacific. The resulting DNA dataset consisted of 6.3 billion base pairs (twice the size of the human genome), with 85% of the assembled and 57% of the unassembled data unique at a 98% identity cutoff. The extreme diversity prevented assembly of complete genomes, as many reads were unique. A comprehensive dataset of GOS sequences combined with pre-exisiting databases reveals nearly 6.12 million proteins, nearly doubling the number of known proteins. Some families of microbial proteins discovered in this study, particularly protein kinases, were previously thought to be restricted to eukaryotic organisms. Over 1700 sequence clusters show no identity to known families, implying we are far from knowing the full range of what proteins can do.
How to make sense of all this data? First, more data is needed!, namely more complete genomes into which the unassembled fragments can be placed. Second, new analytic tools. A new genomics and informatics group based at the California Institute for Telecommunications and Information Technology in San Diego, have built a metagenomics version of GenBank, known as the Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis (try saying that 3 times quickly!) which is fortunately known by acronym CAMERA.
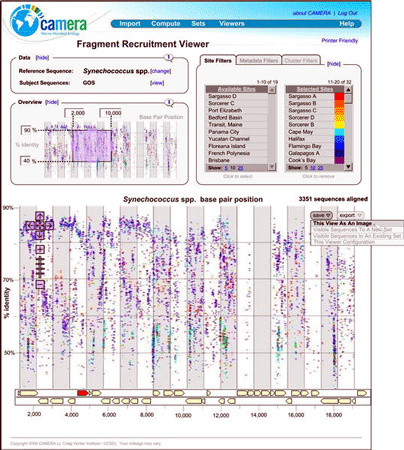
Just as Google and other search engines solved a problem of information overload that did not exist a few years ago, I am confident that CAMERA and other new informatics tools will enable us to view the expanding universe of environmental genomics, including DNA barcode libraries, in ways that will provide new understanding.

 A dozen articles in current issue of
A dozen articles in current issue of 
 The Indomalayan biogeographic region spans a vast area of tropical biodiversity and includes inumerable islands with high numbers of endemic species. A large scale genetic survey with DNA barcoding is likely to help lead to dramatic increases in species counts in particular and better understanding of biodiversity in general. Additional collecting may be particuarly important in this region, as it is at present the least well-represented in frozen tissue collections. There was strong enthusiasm among regional participants, and recognition the initiative has public appeal and the potential to engage new sources governmental support.
The Indomalayan biogeographic region spans a vast area of tropical biodiversity and includes inumerable islands with high numbers of endemic species. A large scale genetic survey with DNA barcoding is likely to help lead to dramatic increases in species counts in particular and better understanding of biodiversity in general. Additional collecting may be particuarly important in this region, as it is at present the least well-represented in frozen tissue collections. There was strong enthusiasm among regional participants, and recognition the initiative has public appeal and the potential to engage new sources governmental support. I look forward to organizational and scientific progress in this exciting region.
I look forward to organizational and scientific progress in this exciting region.