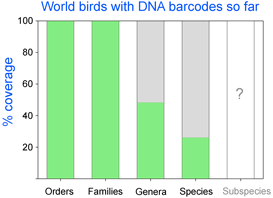
As of October 11, 2008 researchers have deposited 14,594 DNA barcodes in BOLD representing 2,586 avian species, 26% of world’s 9,933 birds. You can browse taxonomic coverage to date at All Birds Barcoding Initiative (ABBI) and BOLD taxonomy browser sites. Coverage includes representatives of all 27 orders and 159 families of world birds and nearly half of avian genera [1,014/2,101 (48%)]. To uncover possible hidden diversity, most researchers are sampling species across their geographic ranges rather than focusing on named subspecies, many or most of which appear to represent clinal variation (see for example Zink 2004, Phillimore and Owens 2006).
As of October 11, 2008 researchers have deposited 14,594 DNA barcodes in BOLD representing 2,586 avian species, 26% of world’s 9,933 birds. You can browse taxonomic coverage to date at All Birds Barcoding Initiative (ABBI) and BOLD taxonomy browser sites. Coverage includes representatives of all 27 orders and 159 families of world birds and nearly half of avian genera [1,014/2,101 (48%)]. To uncover possible hidden diversity, most researchers are sampling species across their geographic ranges rather than focusing on named subspecies, many or most of which appear to represent clinal variation (see for example Zink 2004, Phillimore and Owens 2006).
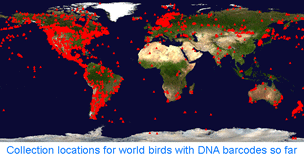
 How far along are researchers toward mapping COI barcode resolution of avian species? Birds are of particular interest because species limits are generally well-defined, supported by a wealth of morphologic, ecological, behavioral, and other genetic data. Looked at regionally, there is good coverage in northern North America, parts of Central and South America, western Europe, Korea, and New Zealand, so it should be possible to see how well COI barcodes distinguish among local species in these areas. Published studies so far show >95% resolution of named species and have identified genetically divergent clusters which may represent unrecognized cryptic or “hidden” species (Vilaca et al 2006, Yoo et al 2006, Nyari 2007, Kerr et al 2007). As an aside, “cryptic” is an awkward term for genetically divergent populations of birds since most of these have diagnostic differences in morphology or behavior; “hidden” is more accurate to my ear. On a separate note, COI surveys have regularly revealed misidentified voucher specimens of birds, suggesting routine application of DNA barcode analysis could enhance quality of avian collections.
How far along are researchers toward mapping COI barcode resolution of avian species? Birds are of particular interest because species limits are generally well-defined, supported by a wealth of morphologic, ecological, behavioral, and other genetic data. Looked at regionally, there is good coverage in northern North America, parts of Central and South America, western Europe, Korea, and New Zealand, so it should be possible to see how well COI barcodes distinguish among local species in these areas. Published studies so far show >95% resolution of named species and have identified genetically divergent clusters which may represent unrecognized cryptic or “hidden” species (Vilaca et al 2006, Yoo et al 2006, Nyari 2007, Kerr et al 2007). As an aside, “cryptic” is an awkward term for genetically divergent populations of birds since most of these have diagnostic differences in morphology or behavior; “hidden” is more accurate to my ear. On a separate note, COI surveys have regularly revealed misidentified voucher specimens of birds, suggesting routine application of DNA barcode analysis could enhance quality of avian collections.
 Looked at globally, there is 100% coverage of 104 polytypic genera (having 2 or more species) representing 324 birds, so this should include the sister species and/or “nearest neighbors” for these, plus there are 853 monotypic genera (having only 1 species) in world birds, which are likely or known to be genetically divergent from birds in other genera. In addition, there are likely many other sister species or “nearest neighbors” within the remaining 1,982 birds with DNA barcodes so far (for example, BOLD includes 28 of 29 Dendroica sp wood warblers). It would be interesting to look at the nearest neighbor differences within the global data set. To my knowledge, comparisons among regions with COI barcode data have been not been published. My impression based on other avian genetic work is that named taxa in different biogeographic regions are genetically distinct, plus there are many unrecognized genetic divisions within species that range across biogeographic regions. I look forward to trans-regional and global comparisons!
Looked at globally, there is 100% coverage of 104 polytypic genera (having 2 or more species) representing 324 birds, so this should include the sister species and/or “nearest neighbors” for these, plus there are 853 monotypic genera (having only 1 species) in world birds, which are likely or known to be genetically divergent from birds in other genera. In addition, there are likely many other sister species or “nearest neighbors” within the remaining 1,982 birds with DNA barcodes so far (for example, BOLD includes 28 of 29 Dendroica sp wood warblers). It would be interesting to look at the nearest neighbor differences within the global data set. To my knowledge, comparisons among regions with COI barcode data have been not been published. My impression based on other avian genetic work is that named taxa in different biogeographic regions are genetically distinct, plus there are many unrecognized genetic divisions within species that range across biogeographic regions. I look forward to trans-regional and global comparisons!
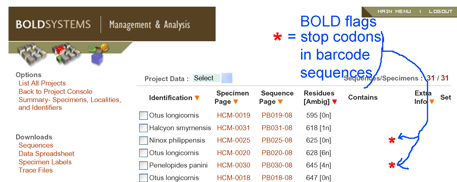
 In
In  In
In  In
In  An article in
An article in  In
In 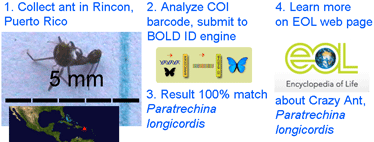 As a test of how DNA barcoding might work for the interested ant novice, I collected the tiny specimen at left in Rincon, Puerto Rico, and submitted its COI barcode to
As a test of how DNA barcoding might work for the interested ant novice, I collected the tiny specimen at left in Rincon, Puerto Rico, and submitted its COI barcode to  Pseudogenes, first described by
Pseudogenes, first described by