Barcoding is a standardized approach to DNA-based species identification. The essence of standardization is an agreement among researchers and practitioners to rely on one or a few defined gene region(s). Standardization makes it possible for researchers to work together to build comprehensive sequence libraries–it is simply not possible for any single group of researchers to collect and analyze the millions of specimens needed to establish a widely-useful reference database. And, looking at the application side, standardization enables species-level identifications without having to know in advance what taxonomic group the specimen belongs to. The standard regions so far are Hebert 2005 COI segment for animals and defined segments of matK and rbcL for land plants. Agreeing on standard barcode regions is a social as well as scientific process–achieving consensus on COI and matK/rbcL are major achievements.
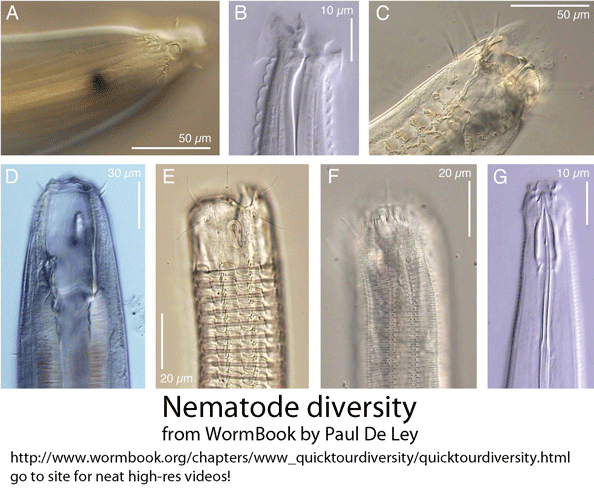
In March 2010 Hydrobiologia, researchers from Indian Institute of Science Education and Research-Kolkata, India, and Plymouth Marine Laboratory, England, report on new primers for amplifying 18S rRNA as a means of barcoding marine nematodes from environmental samples. As background, nematodes are an enormous phylum of mostly tiny and often parasitic worms, including important human, plant, and animal pathogens, and are comprised of many deeply divergent lineages, challenging species-level identification. Despite their ubiquity, diversity, and biological importance, I imagine that most persons are unfamiliar with nematodes. Small subunit (SSU) rRNA (also known as 18S rRNA) is the backbone for nematode molecular phylogeny (Holterman et al 2006). For species-level identification, to my knowledge no single standard has emerged (Blaxter et al 2005, De Ley 2005). SSU/18S rRNA often does not distinguish among species, and so far it has been difficult to reliably amplify COI barcode region from nematodes, presumably due to sequence diversity at primer binding sites. If not COI, then standardizing on a nematode barcode will involve researchers agreeing on defined segment(s), perhaps somewhere in the 7.2 kb ribosomal RNA gene complex.
Back to the paper under discussion–Bhadury and Austen compared two 18S rRNA primer sets: one, previously described (by same authors), which amplifies approximately 345 bp near the 5′ end of 18s rRNA gene, and a new set, which amplifies a 427 bp segment from near the middle of the gene. According to my analysis, these two amplicons, which each represent about 1/5 of 18S rRNA gene, have no overlap. Why select a segment of 18S rRNA as a potential barcode, given that full-length 18S is known to show limited species resolution? In the 2006 paper cited above, the authors explored possible DNA barcoding loci, reporting that “further evaluation with the 28S rRNA, 16S rRNA and COI genes was abandoned as a result of unreliable PCR amplification with several representative marine nematode taxa.” Designing broad-range primers for barcoding nematodes is certainly challenging; this 2006 analysis, based on single specimens of 26 nematode species in 13 families, seems too sparse to make useful conclusions. As to species resolution with 345 bp 5′ 18S fragment, although the abstract states “over 97% of specimens sequenced were correctly assigned,” this turns out to refer to assignments at species OR genus level, and by my reading includes cases that matched to two different genera (with identical 345 bp 18S sequences).
To evaluate the 18S primers, DNA was isolated from two 0.5 g samples of estuary sediment collected in New Jersey, pooled, amplified, and cloned. 60 and 40 clones generated with first and second primer sets, respectively, were sequenced. From first set, 16 haplotypes (comprising 45 clones) showed 89-97% BLAST identity with known nematode sequences and the remaining 4 haplotypes (15 clones) were most similar (88-96% BLAST identify) to non-nematode 18S sequences. This led the researchers to design the second set of primers to reduce co-amplification of non-nematode sequences. The second set produced 6 haplotypes (40 sequenced clones); all were similar or identical (90-100% by BLAST) to published nematode sequences.
Designing primers that selectively amplify barcodes from certain taxa is important in some situations, particularly when analyzing mixtures, such as environmental samples as done here, and also to selectively amplify hosts vs parasites, or ingested DNA in stomach contents vs organism, for example. The authors conclude that “the databases…need to be populated with new full-length 18S rRNA nematode sequences from different biogeographic locations.” More data is always good, but it remains to be seen where efforts should be placed. In nematodes it may be there is a trade-off between having broadly-applicable primers and achieving good species resolution; here more exploration is needed. Agreeing on barcode region(s)s might help lift nematodes, which likely outnumber insects, from obscurity!
Addendum 10 sept 2010 4:10 PM: Dan Janzen points out that there is more to barcoding standards than the above might imply. To whit, the COI barcode is a precisely defined 648-bp segment of COI, and, for inclusion in reference library, barcode sequences need to be accompanied by voucher specimen information, bidirectional trace files with a minimum quality score, and primer sequences.