An excellent article by Sam Moore in the Martha’s Vineyard Times and its magazine Edible Vineyard feature Jesse Ausubel’s views and his collection of eDNA at the Inkwell Beach in Oak Bluffs and elsewhere on the Island.
Best-
An excellent article by Sam Moore in the Martha’s Vineyard Times and its magazine Edible Vineyard feature Jesse Ausubel’s views and his collection of eDNA at the Inkwell Beach in Oak Bluffs and elsewhere on the Island.
Best-
Our environmental DNA discovery of unusual fish is featured in the American Fisheries Society blog. The post highlights the growing complement of East Coast eDNA researchers, including our NOAA colleague Yuan Liu.
Journalist Maurice Tamman wrote an excellent, widely published article, Pandemic offers scientists unprecedented chance to hear oceans as they once were, about the International Quiet Ocean Experiment (IQOE) for Reuters.
NEW YORK (Reuters) – Eleven years ago, environmental scientist Jesse Ausubel dreamed aloud in a commencement speech: What if scientists could record the sounds of the ocean in the days before propeller-driven ships and boats spanned the globe?
even picked up by the New York Times and in the hard copy edition of the Washington Post.
The 2009 Dalhousie University Commencement Address to which it refers is posted here: https://phe.rockefeller.edu/news/2009/11/30/son-et-lumiere-exciting-updates/, published on 23 November 2009 by the monthly science magazine, SEED, as Broadening the Scope of Global Change to Include Illumination and Noise.
The 5th IQOE Newsletter is here.
A vivid set of 2019 articles about the oceans authored by Maurice Tamman and Co.: Maurice Tamman, Matthew Green, Mari Saito, Sarah Slobin and Maryanne Murray – Reuters: Ocean shock: The climate crisis beneath the waves
A good 2019 article in German about ocean sound by Nicola Jones that mentions our work: Das Streben nach leiseren Meeren.
The chief scientist of the Census of Marine Life, Ron O’Dor, passed away in Nova Scotia from COVID-19 at the age of 75 on 11 May 2020. Ron was a curious, good-humored colleague.
Jesse first met Ron in December 1997 at a meeting at the New England Aquarium to assess the feasibility of censusing the “non-fish nekton.” Ron represented the cephalopods, very well. Ron’s creativity immediately became apparent. Chats about the potential of Internet databases with spatial information about marine species led Ron to enlist his grad student James Wood to create CephBase, for all squids and octopi. CephBase launched in 1998, biblical times for on-line services, pre-dating Google. By the time the CoML officially launched at the start of 2000, it was clear the program would need a full-time senior scientist. Ron was the unanimous preference of the founding members of the Scientific Steering Committee.
The CoML went terrifically well in large part because Ron brought many outstanding people into the program, always neatly aligned with CoML objectives. His ability to work on airplanes and in hotel rooms amazed everyone. It did not matter where he was or when it was, work got done, and was always well-written and clean. The Baseline Report that Ron (sole author) produced in October 2003 was a tour de force and did much to establish the credibility of the program.
A remembrance from the Ocean Tracking Network and an obituary in the Chronicle Herald.
The Ron O’Dor Memorial Fund has been set-up to sustain his legacy: giving.dal.ca/ronodor Deep condolences to his widow Janet and family and the Dalhousie science community of which he was a formative member.
National Fisherman May 14, 2020 Genetic markers reveal Brazilian cownose rays, Gulf kingfish in New Jersey waters Kirk Moore
Agencia EFE (via El Diario, Spain) El analísis de ADN medioambiental detecta migraciones de especies marinas https://www.eldiario.es/sociedad/analisis-ADN-medioambiental-migraciones-especies_0_1026448521.html
Prensa Latina, Cuba Descubren nuevos patrones de migración de especies marinas tropicales https://www.prensa-latina.cu/index.php?o=rn&id=365583&SEO=descubren-nuevos-patrones-de-migracion-de-especies-marinas-tropicales
Globedia, Spain Detectan migraciones de especies marinas gracias al ADN medioambiental https://globedia.com/detectan-migraciones-especies-marinas-gracias-adn-medioambiental
Revista Planeta, Brazil Arraia típica do Brasil está chegando perto da costa de Nova York https://www.revistaplaneta.com.br/arraia-tipica-do-brasil-esta-chegando-perto-da-costa-de-nova-york/
DIVE Magazine, UK DNA Traces Prove To Be Useful Tool in Understanding Fish Populations https://divemagazine.co.uk/eco/8973-tracking-and-tracing-in-the-ocean-using-dna-residue
By Jesse H. Ausubel and Vice Admiral Paul Gaffney II. (Ret.), appeared in The Hill 05/07/20
Front. Mar. Sci., 05 May 2020 | https://doi.org/10.3389/fmars.2020.00226
Mark Y. Stoeckle*, Mithun Das Mishu and Zachary Charlop-Powers
An accurate, comprehensive reference sequence library maximizes information gained from environmental DNA (eDNA) metabarcoding of marine fishes. Here, we used a regional checklist and early results from an ongoing eDNA time series to target mid-Atlantic U.S. coastal fishes lacking reference sequences. We obtained 60 specimens representing 31 species from NOAA trawl surveys and institutional collections, and analyzed 12S and COI barcode regions, the latter to confirm specimen identification. Combined with existing GenBank accessions, the enhanced 12S dataset covered most (74%) of 341 fishes on New Jersey State checklist including 95% of those categorized abundant or common. For eDNA time series, we collected water samples approximately twice monthly for 24 months at an ocean and a bay site in New Jersey. Metabarcoding was performed using separate 12S primer sets targeting bony and cartilaginous fishes. Bioinformatic analysis of Illumina MiSeq fastq files with the augmented library yielded exact matches for 90% of the 104 fish amplicon sequence variants generated from field samples. Newly obtained reference sequences revealed two southern U.S. species as relatively common warm season migrants: Gulf kingfish (Menticirrhus littoralis) and Brazilian cownose ray (Rhinoptera brasiliensis). A beach wrack specimen corroborated the local presence of Brazilian cownose ray. Our results highlight the value of strengthening reference libraries and demonstrate that eDNA can help detect range shifts including those of species overlooked by traditional surveys.
We all continue healthy and working long hours and hard, though mostly from our homes. We are catching up on lots of writing and editing but also trying to seize immediate, unique opportunities.
For example, COVID-19 may have created the reduction of additions of human noise that we dreamed about for the International Quiet Ocean Experiment. IQOE welcomes ideas about how the present quieting of the world economy may advance research in marine sound. High-quality observations of the ocean soundscape, as well as possibly related behavior of marine life during this period, may offer unique opportunities of exceptional value.
Resuming our interest in Serious Games, we are also please to encourage a team at the Indian Institute of Technology in Tirupati that is developing SurviveCovid-19 — A Game for Improving Awareness of Social Distancing and Health Measures for Covid-19 Pandemic
Jesse has also written a foreword for Simon Mitton’s forthcoming history of deep carbon science, From Crust to Core, to be published by Cambridge U. Press. https://www.cambridge.org/core/books/from-crust-to-core/E0E2E8FC30B4C784B0FB268AC4AA8371
Dating back to the Ordovician period about 450 million years ago, Bryozoa are small aquatic invertebrates with exoskeletons that typically sieve food particles out of the water with a crown of tentacles. The individual zooids live in colonies forming fans, bushes, and sheets.
Dennis P. Gordon, distinguished taxonomist at New Zealand’s National Institute of Water & Atmospheric Research, has described “New Hippothoidae (Bryozoa) from Australasia” in the journal Zootaxa.
Dennis and Jesse Ausubel worked together in the Census of Marine Life 2000-2010. Dennis has named a new genus of Hippothoidae Bryozoans the Jessethoa and the first species Jessethoa ausubeli.
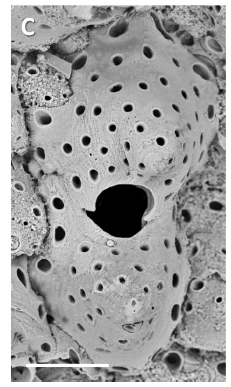
This brings the total of described hippothoid genera to nine (plus two fossil) and species to 83 recent (plus 15 fossil).
The Jessethoa page in the World Registry of Marine Species
The Jessethoa ausubeli page in the World Registry of Marine Species
On behalf of the entire Census of Marine Life, Jesse is greatly honored to be permanently associated with this fascinating taxon. Thank, Dennis Gordon!