 DNA barcodes index species. In most animal species studied so far, mtDNA differences within species are much smaller than those between species. As a result, species appear as distinct clusters in a simple neighbor-joining tree of COI barcodes. The uniformity of this patterning gives confidence that a DNA barcode library based on relatively few individuals per species will be a reliable index for assigning unknown specimens to known species. Although we are just at the beginning of compiling barcodes, and although we need phylogenetically-informed mathematical analysis about how to define clusters particularly in groups not well-studied, I am struck by how obvious most species clusters are. There are of course exceptions and limits (hybridization, young species, slow mitochondrial DNA evolution) but it is likely that someone with no knowledge other than a neighbor-joining tree of DNA barcodes could reconstruct most species categories, although they wouldn’t know anything about the biology of the organisms. This suggests viewing DNA barcoding as a diagnostic tool that links to biological knowledge, just as a laboratory test is used to detect HIV for example, and thereby point to a large body of biological knowledge.
DNA barcodes index species. In most animal species studied so far, mtDNA differences within species are much smaller than those between species. As a result, species appear as distinct clusters in a simple neighbor-joining tree of COI barcodes. The uniformity of this patterning gives confidence that a DNA barcode library based on relatively few individuals per species will be a reliable index for assigning unknown specimens to known species. Although we are just at the beginning of compiling barcodes, and although we need phylogenetically-informed mathematical analysis about how to define clusters particularly in groups not well-studied, I am struck by how obvious most species clusters are. There are of course exceptions and limits (hybridization, young species, slow mitochondrial DNA evolution) but it is likely that someone with no knowledge other than a neighbor-joining tree of DNA barcodes could reconstruct most species categories, although they wouldn’t know anything about the biology of the organisms. This suggests viewing DNA barcoding as a diagnostic tool that links to biological knowledge, just as a laboratory test is used to detect HIV for example, and thereby point to a large body of biological knowledge.
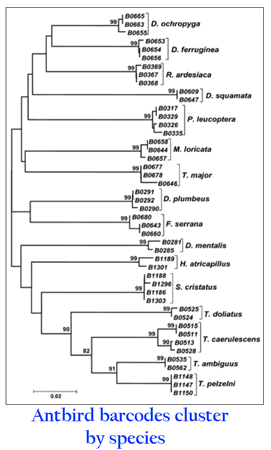
There are more bird species in the Neotropics than anywhere else. Over 4,000 of the approximately 10,000 world bird species live in South and Central America and the Caribbean, including over 3000 endemics. The large number of speciose families and the fact that intraspecific genetic variation is generally thought to be greater in the tropics than in temperate regions (eg Balakrishnan 2005 Syst Biol 54:689) might challenge DNA barcoding. In what I believe is the first explicit application of DNA barcoding to Neotropical birds (Vilaca 2006 Revista Brasileira Ornitologia 14:7) researchers analyzed 16 species of antbirds in the Atlantic Forest region of southeastern Brazil, with half of specimens obtained as blood samples from birds in the field. All species form distinct clusters in a neighbor-joining tree with 99% bootstrap support, including the recently split pair Thamnophilus pelzelni (shown above) and T. ambiguus. Maximum intraspecific variation is less than 1% except in T. caerulescens which shows 2 distinct lineages, highlighting a good candidate for further study.
 Do you see a new species anywhere? Baleen whale specimen collected in 1976, new species description 27 years later based in part on mitochondrial DNA characters (Wada et al. 2003. Nature 426:278)
Do you see a new species anywhere? Baleen whale specimen collected in 1976, new species description 27 years later based in part on mitochondrial DNA characters (Wada et al. 2003. Nature 426:278) 
 With funding from the Gordon and Betty Moore Foundation, researchers at Reveo, Inc. and the University of Washington are collaborating on developing a
With funding from the Gordon and Betty Moore Foundation, researchers at Reveo, Inc. and the University of Washington are collaborating on developing a 
 Brower’s article argues about how many Astraptes species are supported by the sequence data, worries about possible consequences of the rise of DNA barcoding, and misses the scientific opportunity to examine this very young species complex with a large set of morphologic, ecologic, and sequence data. He concedes “there are probably at least three species” but declines to put forth what criteria can be used to delimit species. I was surprised to read that the question of whether two sequences “belong to the ‘same species’ is a metaphysical one”.
Brower’s article argues about how many Astraptes species are supported by the sequence data, worries about possible consequences of the rise of DNA barcoding, and misses the scientific opportunity to examine this very young species complex with a large set of morphologic, ecologic, and sequence data. He concedes “there are probably at least three species” but declines to put forth what criteria can be used to delimit species. I was surprised to read that the question of whether two sequences “belong to the ‘same species’ is a metaphysical one”. 

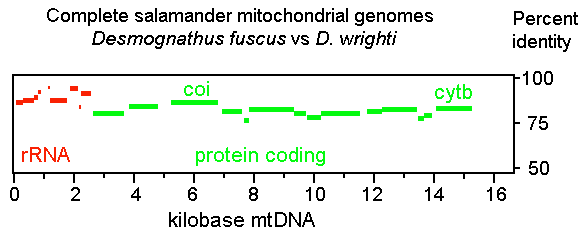
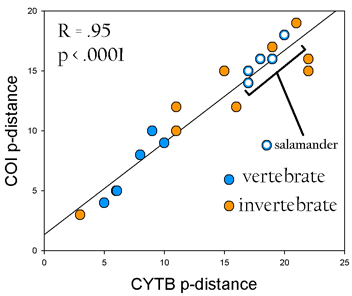







 A year ago in Science,
A year ago in Science,