Approximately 8,000 – 15,000 species of bivalves (clams, mussels, scallops, oysters, and relatives) are known. According to BOLD Taxonomy Browser www.barcodinglife.org, 620 bivalve species have COI barcode records so far, so this group is relatively unexplored genetically. In September 2007 Zoologica Scripta researchers from University of Bergen, Norway, analyze COI barcode region sequences of 62 deepwater clams dredged in a single offshore region at 69 m to 567 m, morphologically identified as 12 species from 4 genera (Thyasira, Ennucula, Nucula, Yoldiella) representing 3 subclasses of Bivalvia. The COI barcode region was amplified with broad-range primers (Folmer et al 1994). Mean differences within species collected in this single area were small, 0.0 – 0.48%, similar to results in other animal groups, suggesting assignment of specimens to species will be straightforward. This will be helpful in environmental surveys for example, as some species “are infamous for being difficult to determine to species from morphology” and some “remain difficult to identify for the non-expert.” As one example, some Thyasira species are distinguished only by sperm and egg morphology, which is impractical in most circumstances.
 mtDNA differences among these bivalves are remarkably large, even among species in the same genus. The differences among congeneric species in this sample (average 22%, range 12-42%) are larger than differences among entire class Aves (according to my analysis with BOLD software, COI differences among birds in different orders, such as penguins and hummingbirds for example, average 20%, with range 14-28%).
mtDNA differences among these bivalves are remarkably large, even among species in the same genus. The differences among congeneric species in this sample (average 22%, range 12-42%) are larger than differences among entire class Aves (according to my analysis with BOLD software, COI differences among birds in different orders, such as penguins and hummingbirds for example, average 20%, with range 14-28%).
Blastn GenBank searches with these divergent mtDNA sequences showed very limited identity to anything, and the closest matches were short stretches (100-150 nucleotides of the 678 full-length barcode sequence) to COI sequences of species outside the phylum Mollusca (I obtained similar results submitting Thyasira sequences for example to the public BOLD Identification Engine at www.barcodinglife.org.) It will be helpful if Mikkelsen et al deposit their sequences along with associated collecting data (voucher specimen information, images, collection locations) to the BOLD database. I look forward to learning more about these bivalves, and whether their remarkably deep differences in mtDNA are associated with deep physiological, ecological, or other biological differences.
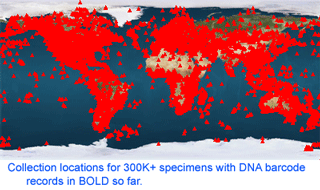 As of 28 january 2008, there are 341,825 barcode records from 35,798 species in the Barcode of Life Database (BOLD)
As of 28 january 2008, there are 341,825 barcode records from 35,798 species in the Barcode of Life Database (BOLD) 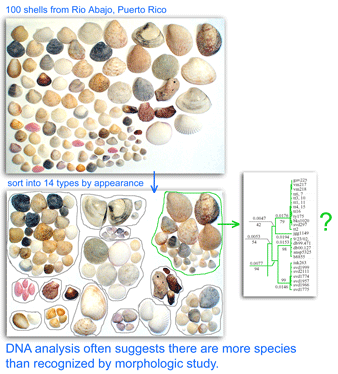 One result is that it has been difficult to compare patterns of diversification between different branches in the Tree. One might ask, how finely and how evenly divided is biodiversity? Are the differences among and within mosquito species (3,400 species), for example, similar to the differences among and within fruit flies (6,200 species) or birds (10,000 species)? Broad application of DNA analysis is beginning to provide some insights. To enable these sorts of comparisons, a standardized locus is needed, as unique genes can solve local branching patterns, but do not allow easy comparisons between branches.
One result is that it has been difficult to compare patterns of diversification between different branches in the Tree. One might ask, how finely and how evenly divided is biodiversity? Are the differences among and within mosquito species (3,400 species), for example, similar to the differences among and within fruit flies (6,200 species) or birds (10,000 species)? Broad application of DNA analysis is beginning to provide some insights. To enable these sorts of comparisons, a standardized locus is needed, as unique genes can solve local branching patterns, but do not allow easy comparisons between branches.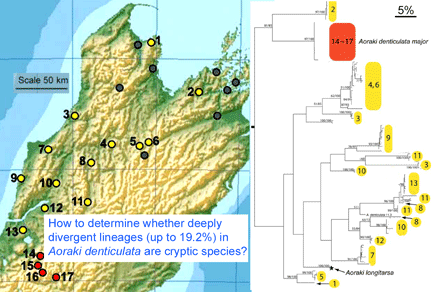
 Lumpsuckers are globular, scaleless marine fish with bony tubercles on head and body, and a ventral sucking disc, derived from specialized pelvic fins, which allows them to adhere to environmental substrates. The genus Eumicrotremus comprises 16 species distributed in the Arctic and northern Atlantic and Pacific oceans; the commonest and most widespread in the north Atlantic is the Spiny lumpsucker E. spinosus, which was first described by Fabricius in 1776. A new subspecies E. s. eggvinii was described in 1956, based on a single specimen, and this was later elevated to species level “on the basis of wrinkled skin, numerous dermal warts and a large sucking disk, in addition to the low number of bony tubercles.”
Lumpsuckers are globular, scaleless marine fish with bony tubercles on head and body, and a ventral sucking disc, derived from specialized pelvic fins, which allows them to adhere to environmental substrates. The genus Eumicrotremus comprises 16 species distributed in the Arctic and northern Atlantic and Pacific oceans; the commonest and most widespread in the north Atlantic is the Spiny lumpsucker E. spinosus, which was first described by Fabricius in 1776. A new subspecies E. s. eggvinii was described in 1956, based on a single specimen, and this was later elevated to species level “on the basis of wrinkled skin, numerous dermal warts and a large sucking disk, in addition to the low number of bony tubercles.”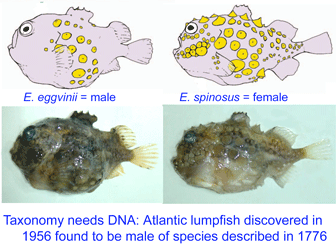 In August 2007
In August 2007  In 17 September 2007 Zootaxa (
In 17 September 2007 Zootaxa (