The lead story in the US National Museum of Natural History 2005 Annual Report: New Tools for Understanding Nature, entitled “Barcoding the Planet” highlights the  museum’s organizational and research involvement in the international scientific effort “to develop a system for rapidly and inexpensively identifying the approximately 1.7 million known flora and fauna species, and creating an electronic database for the estimated 10 million species across the planet”. As outlined by Cristian Samper, Museum Director, “the use of DNA barcoding in identifying and distinguishing species could revolutionize the way we do science”. The article concludes with an observation from Lee Weigt, Manager of the Museum’s Laboratories of Analytical Biology “What the human genome research can do for medicine, DNA barcoding can do for biology”
museum’s organizational and research involvement in the international scientific effort “to develop a system for rapidly and inexpensively identifying the approximately 1.7 million known flora and fauna species, and creating an electronic database for the estimated 10 million species across the planet”. As outlined by Cristian Samper, Museum Director, “the use of DNA barcoding in identifying and distinguishing species could revolutionize the way we do science”. The article concludes with an observation from Lee Weigt, Manager of the Museum’s Laboratories of Analytical Biology “What the human genome research can do for medicine, DNA barcoding can do for biology”
News
DNA barcoding identifies mystery hummingbird, points toward wide utility in conservation assessments
An unidentified Selasphorus hummingbird spent fall 2005 and winter 2006 frequenting a hummingbird feeder in London, Ontario. As is often true with female or immature hummingbirds, despite close observation and photographs,  it was not possible to identify the exact species, in this case whether this was an Allen’s (S. sasin) or Rufous (S. rufus), species native to the western U.S. that normally winter in Mexico. Even in the hand, identification can be difficult and in banding studies most individuals are often simply recorded as “UNHU”, unidentified hummingbird species.
it was not possible to identify the exact species, in this case whether this was an Allen’s (S. sasin) or Rufous (S. rufus), species native to the western U.S. that normally winter in Mexico. Even in the hand, identification can be difficult and in banding studies most individuals are often simply recorded as “UNHU”, unidentified hummingbird species.
In this case, a single feather  spotted beneath the feeder was brought to University of Guelph, Ontario. DNA extracted from the feather and analyzed for COI barcode proved a match for S. rufus.
spotted beneath the feeder was brought to University of Guelph, Ontario. DNA extracted from the feather and analyzed for COI barcode proved a match for S. rufus.
Beyond solving a conundrum for birders, this case points toward a general utility of DNA barcoding in conservation assessments by enabling routine identification of otherwise unidentifiable species, including use of samples from live individuals which may be particularly important in study of threatened or endangered species.
Palearctic birds barcoding workshop held in Netherlands
Researchers met at Naturalis, the Natural History Museum of Leiden, Netherlands, on 20-21 April 2006 to form plans for barcoding Palearctic birds. The meeting was convened by Per Ericson,  ABBI Palearctic Regional Chair, Swedish Museum of Natural History, hosted by Rene Dekker, Naturalis, and included representatives from Canada, Denmark, England, France, Iran, Italy, Japan, Norway, Portugal, South Korea, Sweden, and the United States. Additional participants in Palearctic ABBI are welcome–please contact Per Ericson per.ericson@nrm.se.
ABBI Palearctic Regional Chair, Swedish Museum of Natural History, hosted by Rene Dekker, Naturalis, and included representatives from Canada, Denmark, England, France, Iran, Italy, Japan, Norway, Portugal, South Korea, Sweden, and the United States. Additional participants in Palearctic ABBI are welcome–please contact Per Ericson per.ericson@nrm.se.
Discussion topics included sampling strategies based on geographical patterns in Palearctic avian diversity, updating the ABBI compilation of existing avian tissue specimens, using ongoing collecting and ringing operations as additional sources for un- or under-sampled species, cost-effectiveness of DNA sequence recovery from museum skins, facilitation of regional network activities with Barcode of Life Database (BOLD), need for email, website, and/or listserve to monitor progress and plan next steps, potential small and large-scale funding sources including possible low or no-cost sequencing at pre-existing genomic centers, and exciting early results with 600+ COI barcodes from eastern Palearctic birds. Based on this workshop, the Palearctic group expects much progress over the coming year.
Barcode Blog
With DNA barcoding for species identification creating excitement and controversy, Mark Stoeckle has launched the “Barcode Blog†to share the community news.
Sometimes taxonomy moves slowly, could use help
In 1998, as part of biodiversity survey to assess the health of New York City’s Central Park, researchers at the American Museum of Natural History collected leaf litter samples. After sorting, the museum sent a collection of Central Park millipedes and centipedes to Richard L. Hoffman, curator for invertebrates at the Virginia  Museum of Natural History. After study, Dr. Hoffman sent several specimens he could not identify to scientists in Italy. In July 2002, Italian scientists announced the Central Park centipede was a new species, and named it Nannarrup hoffmani in honor of Dr. Hoffman. In October 2003, Foddai, Bonato, Pereira, and Minelli published the species description in Journal of Natural History. As of April 2006, the journal issue containing the description is available to subscribers or by payment per article.
Museum of Natural History. After study, Dr. Hoffman sent several specimens he could not identify to scientists in Italy. In July 2002, Italian scientists announced the Central Park centipede was a new species, and named it Nannarrup hoffmani in honor of Dr. Hoffman. In October 2003, Foddai, Bonato, Pereira, and Minelli published the species description in Journal of Natural History. As of April 2006, the journal issue containing the description is available to subscribers or by payment per article.
My summary: 1 new species, found across the street from one of the world’s premier natural history research institutions, recognized as a new species 4 years later, published description year 5, awaiting public access year 8. I believe that DNA barcoding can provide taxonomists with scientific tools and help attract funding to accelerate this process.
DNA helps save sharks
Many shark species are threatened by overfishing, including the  filter-feeding whale shark Rhincodon typus. The Ocean Conservancy reports “many sharks fall victim to finning, the process of slicing off a shark’s fins and tossing it back into the water. Highly prized for use in the delicacy shark fin soup, shark fins support a very lucrative market. Although the U.S. Shark Finning Prohibition Act of 2000 made this practice illegal in all U.S. waters, finning remains legal
filter-feeding whale shark Rhincodon typus. The Ocean Conservancy reports “many sharks fall victim to finning, the process of slicing off a shark’s fins and tossing it back into the water. Highly prized for use in the delicacy shark fin soup, shark fins support a very lucrative market. Although the U.S. Shark Finning Prohibition Act of 2000 made this practice illegal in all U.S. waters, finning remains legal  in most parts of the world.” In The USA regulations prohibit possession of any part of protected species, but how to identify dried fins? The National Oceanographic and Atmospheric Administration recently confiscated a ton of dried shark fins and brought charges against dealers based on DNA identification of protected shark species. A DNA barcode library together with rapid, portable methods for sequence analysis will empower enforcement of regulations for many protected species.
in most parts of the world.” In The USA regulations prohibit possession of any part of protected species, but how to identify dried fins? The National Oceanographic and Atmospheric Administration recently confiscated a ton of dried shark fins and brought charges against dealers based on DNA identification of protected shark species. A DNA barcode library together with rapid, portable methods for sequence analysis will empower enforcement of regulations for many protected species.
Barcoding aids biodiversity science, new examples; some taxonomists worry, part 2
DNA barcoding helps conserve biodiversity. In Ten Reasons for Barcoding Life we outlined benefits to biodiversity from barcoding. An earlier post highlighted one reason, namely how barcoding assists conservation by helping uncover cryptic species. I highlight two more with new examples, and discuss a recent worry piece on barcoding from Conservation Science. From Ten Reasons:
1. Identifying a species from bits and pieces. Conserving the many threatened fish species requires the ability to identify commercial and sport fishery harvests. The United States Food and Drug Administration online Regulatory Fish Encyclopedia was developed to help federal, state, and local officials and purchasers of seafood accurately identify species substitution and economic deception in the marketplace. The page for the Caribbean species Red Snapper Lutjanus campechanus includes high-resolution pictures of filets. However, it is not possible to accurately identify fish fillets and substitution appears common. Using mitochondrial DNA (cytochrome c rather than cytochrome oxidase I), Marko et al found that 77% of Red  Snapper filets sold in eastern USA consumer markets were mislabelled. More than half of the analysed sequences grouped closely with species from other regions of the world. A reference library of DNA barcodes will help regulatory agencies enforce fish quotas and may enable new forms of certification that will be of great interest to the many concerned consumers of regulated products.
Snapper filets sold in eastern USA consumer markets were mislabelled. More than half of the analysed sequences grouped closely with species from other regions of the world. A reference library of DNA barcodes will help regulatory agencies enforce fish quotas and may enable new forms of certification that will be of great interest to the many concerned consumers of regulated products.
What fish is this?

2. Makes expertise go further. Mosquito control programs depend on accurate species identification, a task requiring great expertise, particuarly for larval forms, which can provide early warning before adults hatch and can be treated with local measures. Once a reference library of DNA barcodes is established, DNA-based identification can be applied by many more personnel to more effectively target control measures and limit injury to non-harmful species.
To spray or not to spray?
Worried taxonomists (part 2). In April 2006 Conservation Biology, Daniel Rubinoff’s mildly-titled essay “The utility of mitochondrial DNA barcodes in species conservation” focuses on the potential consequences of relying on DNA barcodes IF they were to be used as the sole criterion for species discovery. Before commenting on this issue, I point out the essay leaves out the multiple practical benefits to biodiversity science which arise from allowing many more people to identify the species around them. As things stand at present, outside of a few well-known groups such as birds and butterflies, taxonomic identification of recognized species is the province of a few experts. Even an expert can identify only a small part of the plant and animal kingdoms, and often cannot identify all stages of life because they lack morphologic characters. A reference library of DNA barcodes will assist experts as well as users. The Rubinoff essay raises interesting and important issues, but they are NOT arguments against the aims of DNA barcoding as envisioned by most practitioners. As outlined on the Consortium for the Barcode of Life (CBOL) website, DNA barcoding is being developed as a tool for taxonomic science, not a replacement for it. The Rubinoff essay does allow that “for groups that are already relatively well known, especially birds and mammals, molecular studies based on barcode-sized sequences have revealed cryptic DNA lineages and may be helpful”. However, the essay states that “the use of DNA to survey already studied groups or test hypotheses…is not truly barcoding”. Rubinoff provides his own definition: “true barcoding consists of broad, essentially blind and random surveys of communities with little or no background information,” which certainly sounds like a scary idea! If analyzing known groups is not barcoding, this will be news to CBOL, as the current initatives include include All Fishes, All Birds, known commercially-important species of Canada, known pests and invasive species, moths of North America, and sphinx and saturnid moths of the world (see CBOL website). (For an idea of the range of activities sparked by the CBOL intiative, see the published symposium from the First International Conference on Barcoding Life held at the Natural History Museum, London, 7-9 February 2005.) It is likely that these large scale surveys of known groups will uncover cryptic lineages, as Rubinoff describes, but they are far from “essentially blind and random surveys of groups with little or no background information”.
It is from large-scale analyses of recognized species in well-studied groups that we will be able to develop confidence in and understand the limitations to barcoding as a tool for species identification and discovery. That said, it is an unremarkable observation that high levels of mitochondrial divergence often signal new species. Ultimately it may be possible to apply confidence thresholds to various levels of mitochondrial sequence divergence, as Chris Meyer and Gustav Paulay’s work on cowries suggests, but this will require much more data gathering. The issue of what are the best quantatitives measures of biodiversity is contentious, and DNA sequences are certain to only be a part of that.
For the next critical essay on DNA barcoding, I offer the writer(s) the following observations:
1. DNA barcoding is a taxonomic tool for a) assigning specimens to known species and b) speeding discovery of new species. More work is needed to determine the best use of DNA barcodes in species discovery (for example, distance vs. character-based methods).
2. Barcode sequences may be of interest to those studying deep phylogeny, but barcoding does not aim to analyze evolutionary groupings above the species level.
3. In some cases a COI barcode narrows identification to a few closely-related species, but no further.
4. Divergent sequence clusters lacking biological co-variants (eg morphologic characters) signal a need for further taxonomic study.
I close with an inspiring quote from an in press Royal Proceedings article by Paul Barber and Sarah Boyce applying DNA barcoding to analyze diversity in stomatopod larvae. They call for an “iterative process of DNA barcoding…followed by taxonomic study…Such a synergy between molecular geneticists and taxonomists will greatly advance our understanding, description and cataloguing of our planet’s biodiversity, moving us closer to the goal of documenting the entirety of the world’s biodiversity in both marine and terresterial environments. However, this synergy will only be possible if funding is directed both towards barcoding efforts as well as traditional morphological-based taxonomy that successful barcoding efforts will require.”
“Taxonomy for the twenty-first century” and “DNA barcoding of life”: special theme issues of Philosophical Transactions of the Royal Society available online

“Taxonomy for the twenty-first century” the 29 April 2004 special theme issue of Philosophical Transactions of the Royal Society Biological Sciences is available online. Allen Herre recommends this compilation as “an extremely useful source of information and ideas on what taxonomy is, what its needs are (and our needs of it), and where it is going.”
Introduction by H. C. J. Godfray and S. Knapp
Taxonomic triage and the poverty of phylogeny by Quentin D. Wheeler
A taxonomic wish-list for community ecology by Nicholas J. Gotelli
Protist taxonomy: an ecological perspective by Bland J. Finlay
Stability or stasis in the names of organisms: the evolving codes of nomenclature by Sandra Knapp, Gerardo Lamas, Eimear Nic Lughadha, et al.
Prokaryote diversity and taxonomy: current status and future challenges by Aharon Oren
Taxonomy and fossils: a critical appraisal by Peter L. Forey, Richard A. Fortey, Paul Kenrick, et al.
Automated species identification: why not? by Kevin J. Gaston and Mark A. O’Neill
The promise of a DNA taxonomy by Mark L. Blaxter
Towards a working list of all known plant species by Eimear Nic Lughadha
Biodiversity informatics: managing and applying primary biodiversity data by Jorge Soberón and Townsend Peterson
Unitary or unified taxonomy? by Malcolm J. Scoble
The role of taxonomy in species conservation by Georgina M. Mace
Taxonomy and environmental policy by Cristián Samper
Taxonomy: where are we now? by Peter H. Raven
Now is the time: by Daniel H. Janzen
Tomorrow’s taxonomy: collecting new species in the field will remain the rate-limiting step by Robert M. May
Documenting plant diversity: unfinished business by Peter R. Crane
Taxonomy as a fundamental discipline by Edward O. Wilson
“DNA barcoding of life” the 29 October 2005 issue of Philosophical Transactions of the Royal Society Biological Sciences is devoted to papers presented at the First International Barcode Conference held at The Natural History Museum, London, 7-9 February 2005, a gathering that included 220 participants from 44 countries. By special arrangement with The Royal Society, this issue is available to members and visitors to the Consortium of the Barcode of Life site.
Towards writing the encyclopaedia of life: an introduction to DNA barcoding by Vincent Savolainen, Robyn S. Cowan, Alfried P. Vogler, et al.
DNA barcodes for biosecurity: invasive species identification by K.F. Armstrong and S.L. Ball
DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar by M. Alex Smith, Brian L. Fisher, Paul D.N. Hebert
Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding by Daniel H. Janzen, Mehrdad Hajibabaei, John M. Burns, et al.
DNA barcoding Australia’s fish species by Robert D. Ward, Tyler S. Zemlak, Bronwyn H. Innes, et al.
Deciphering amphibian diversity through DNA barcoding: chances and challenges by Miguel Vences, Meike Thomas, Ronald M. Bonett, et al.
The problems and promise of DNA barcodes for species diagnosis of primate biomaterials by Joseph G. Lorenz, Whitney E. Jackson, Jeanne C. Beck, et al.
Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications by Gary W. Saunders
Land plants and DNA barcodes: short-term and long-term goals by Mark W. Chase, Nicolas Salamin, Mike Wilkinson, et al.
Microcoding: the second step in DNA barcoding by R.C. Summerbell, C.A. Lévesque, K.A. Seifert, et al.
The unholy trinity: taxonomy, species delimitation and DNA barcoding by Rob DeSalle, Mary G. Egan, Mark Siddall
Reverse taxonomy: an approach towards determining the diversity of meiobenthic organisms based on ribosomal RNA signature sequences by Melanie Markmann and Diethard Tautz
DNA-based species delineation in tropical beetles using mitochondrial and nuclear markers by Michael T. Monaghan, Michael Balke, T. Ryan Gregory, et al.
Defining operational taxonomic units using DNA barcode data by Mark Blaxter, Jenna Mann, Tom Chapman, et al.
An integrated approach to fast and informative morphological vouchering of nematodes for applications in molecular barcoding by Paul De Ley, Irma Tandingan De Ley, Krystalynne Morris, et al.
Critical factors for assembling a high volume of DNA barcodes by Mehrdad Hajibabaei, Jeremy R. deWaard, Natalia V. Ivanova, et al.
A likelihood ratio test for species membership based on DNA sequence data by Mikhail V. Matz and Rasmus Nielsen
TaxI: a software tool for DNA barcoding using distance methods by Dirk Steinke, Miguel Vences, Walter Salzburger, et al.
Some Fret Over Exceptions to Barcoding
The springboard for a recent news@nature.com item by Hannah Hickey “Butterflies poke holes in DNA barcodes” is a report by Gompert et al in press in Mol. Ecology on genetic differences between two subspecies of Melissa blue butterfly, Lycaeides melissa melissa and L. m. samuelis. The latter subspecies is commonly known as “Karner blue” and is listed under the USA Endangered Species Act. Analysis of mitochondrial DNA revealed some populations of Karner blue have distinct COI sequences but those populations adjacent to the range of L. m. melissa subspecies do not. This result is not surprising. For one, DNA barcoding does not aim to separate subspecies. Subspecies are geographic variants within species whose differences shade into one another so it would be surprising if any single gene showed a sharp demarcation between populations. Most subspecies do not show diagnostic genetic differences, and when such differences are found, it has often led to proposals to elevate them to species status.
Regarding the utility of DNA barcoding, the findings with Melissa blues are unremarkable, as there are cases in all animal groups studied so far in which barcoding narrows identification to a few closely-related species, but no further. For example, see my earlier entry on comparing barcode performance. It may be helpful to point out that DNA barcoding is an instrument, not a theory. Cases of partial resolution do not “disprove” barcoding or invalidate its use. In fact, one application of DNA barcoding will be to quickly highlight such cases which may be biologically interesting as they likely represent recent speciation, ongoing hybridization, or synonymy.
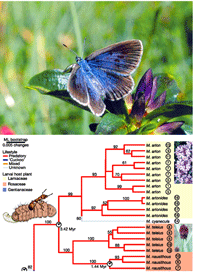
A more relevant Nature article that Ms. Hickey might have cited is Als et al study of Maculinea large blues, a related group morphologically similar, taxonomically confusing, and highly endangered butterflies. Large blues have “extraordinary parasitic lifestyles…later instars live in ant nests where they either devour the brood (predators), or are fed mouth-to-mouth by adult ants (cuckoos)”. Genetic analysis using mitochondrial and nuclear DNA uncovered numerous cryptic species with unsuspected host specificity, thereby both multiplying the challenge and providing the key to conservation, the need to conserve both ant hosts and butterflies.
As highlighted by the large blue study, the larger and more exciting challenge for biodiversity science will be how to incorporate the enormous number of genetically and biologically distinct forms whose discovery is facilitated by large-scale barcoding.
Photo of Rebel’s large blue Maculinea rebeli and phylogeny showing cryptic species among predatory Maculinea from Nature article by Als et al.
Jacques Perrin
The festival in Washington DC celebrating the nature films of Jacques Perrin opened last night with an event highlighting Galatee’s new Oceans Project. We post Jesse’s Welcoming Remarks to Jacques Perrin.