 Insiders can be mistaken, in science and in other fields. At the beginning of the Human Genome Project, “the great majority of scientists dismissed the original proposal with hostility or indifference” (Great 15-year project to decipher genes stirs opposition. New York Times, June 5, 1990). The Times article details some of the initial negative reactions:
Insiders can be mistaken, in science and in other fields. At the beginning of the Human Genome Project, “the great majority of scientists dismissed the original proposal with hostility or indifference” (Great 15-year project to decipher genes stirs opposition. New York Times, June 5, 1990). The Times article details some of the initial negative reactions:
“Even if scientists manage to finish the genome project, it will have generated enormous reams of uninterpretable and often useless data”.
“The human genome project is bad science, it’s unthought-out science, it’s hyped science” said Dr. Martin Rechsteiner, a biochemist at the University of Utah. Some critics have begun aggressive letter-writing campaigns, urging colleagues who harbor similar sentiments to write Congress.
“Everybody I talk to thinks this is an incredibly bad idea,” said Dr. Michael Syvanen, a microbiologist at the Medical School of the University of California at Davis and a stout antagonist of the genome project.
Professional societies weighed in as well. A resolution adopted by the Council of the American Society for Biochemistry and Molecular Biology, and endorsed by the Federation of American Societies for Experimental Biology stated: “A large scale, massive effort to ascertain the sequence of the entire genome cannot be adequately justified at the present time… The Council wants to state in the clearest possible terms our opposition to any current proposal that envisions the establishment of one or a few large centers that are designed to map and/or sequence the human genome.” https://www.fasebj.org/cgi/reprint/1/6/502
This history comes to mind in reading the article by Hickerson, Meyer, and Moritz in October 2006 Syst Biol 55:729. According to their analysis, mathematical modelling predicts that DNA barcoding will often fail to discover young species. Their analysis is based on a classical model of speciation (Bateson-Dobzhansky-Muller) and “well-established population genetic theory”. I should tread lightly here, not being a population biologist! To my reading, these mathematical models are either unsupported or disproved by experimental evidence. The BDM model of biological species formation is “well-characterized, tractable, and its dynamics captures a range of speciation times implicit across many pre- and post-zygotic isolation models”, ie good for modelling, but is not derived from actual genetic data on differences between sister species. Genetic surveys including growing barcode libraries demonstrating limited intraspecific variation in diverse species across enormous differences in population size and generation time indicate that “well established population genetic theory” does not explain intraspecific mitochondrial diversity (Bazin et al 2006 Science 28:570).
Instead of making predictions about why barcoding will fail, I hope the same mathematic rigor will be applied to understanding why barcoding works as well as it does, why the variation within most species is low, why the distances between most species are large, and what determines the exceptions.
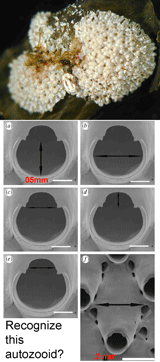

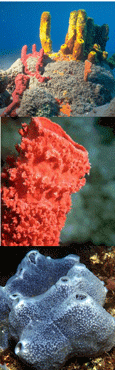 The Sponge Barcoding Project
The Sponge Barcoding Project 
 In 1987, the few dozen GPS models available were mostly larger than 200 cu in and cost $15,000 to $45,000. (
In 1987, the few dozen GPS models available were mostly larger than 200 cu in and cost $15,000 to $45,000. ( Good news for taxonomic science:
Good news for taxonomic science: 
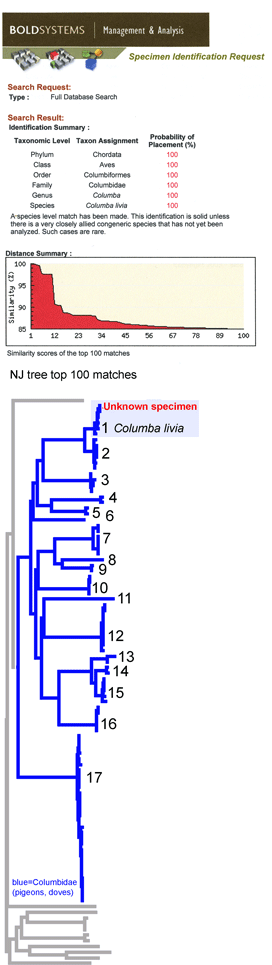 This simulation tries out what barcode identification might be like once reference libraries are established, and corresponds to “species identification” (vs species discovery) in last week’s post. A sequence was selected from Barcodes of Life Data Systems (BOLD) (130,000 COI barcode sequences from 19,000 species so far) and pasted into public “Identification Engine” on
This simulation tries out what barcode identification might be like once reference libraries are established, and corresponds to “species identification” (vs species discovery) in last week’s post. A sequence was selected from Barcodes of Life Data Systems (BOLD) (130,000 COI barcode sequences from 19,000 species so far) and pasted into public “Identification Engine” on 