Jesse becomes engaged in the Creationism discussion, when asked
whether all life might actually voyage on Noah’s Ark, as reported
in an article Reuters’ excellent environment
correspondent, Alister Doyle.
FEATURE-How did Noah’s Ark float? New species cram aboard
Thu May 15, 2008 8:04am EDT
By Alister Doyle, Environment Correspondent
OSLO, May 15 (Reuters) – How did Noah’s Ark manage to stay afloat?
Estimates of the number of species on earth are surging into apparently hull-busting millions as biologists find new life almost everywhere they look, from African swamps to Antarctica.
The ever-widening menagerie is a paradox when an expanding human population, pollution and climate change threaten what United Nations’ studies say is the worst spate of extinctions since the dinosaurs were wiped out 65 million years ago.
Government officials trying to protect the modern world’s wildlife gather in Bonn from May 19-30 for a meeting of the U.N. Convention on Biological Diversity, to examine progress towards a goal set in 2002 of slowing biodiversity loss by 2010.
Most experts say the target is slipping out of reach.
Even so, wider research means finds of new species such as a legless lizard in Brazil or a Tanzanian shrew are testing biblical scholars’ calculations on how Noah squeezed all animals aboard the Ark.
“It’s of course physically impossible,” James Edwards, executive director of the Encyclopedia of Life, said of the biblical account.
The Encylopedia is cataloguing all identified species, 1.8 million so far, in a free online service (https://www.eol.org/).
“There are expectations of 8 to 50 million more species out there that we haven’t identified yet,” Edwards said. Other experts’ estimates of the numbers range up to 100 million.
But the newly found species do not compensate for extinctions.
Sigmar Gabriel, environment minister for the U.N. conference host Germany, said last week that the loss of species threatened food supplies for billions of people. He cited marine life, saying that if nothing was done there would be no commercial fishing by 2050.
Extinctions of recent decades include Australia’s southern gastric brooding frog — the females could shut off their stomach juices to raise young in their stomachs, a trick that could have held clues for curing human ulcers.
Believers in the Bible note that the Ark described in the Book of Genesis was a giant ship by ancient standards about 140 metres (460 ft) long — far from the tiny vessel depicted in many children’s books with giraffes’ heads sticking out the top.
In the biblical account, Noah safeguarded life on the planet after God, upset by the wickedness and violence of mankind, sent a devastating flood. “Everything on earth will perish,” God said, according to the Bible.
ALL CREATURES
Noah only took along land creatures and birds, not plants nor fish that make up a large part of the world’s total species.
“We’re talking about something plausible here,” said David Menton, an associate professor emeritus of anatomy at Washington University who works for Answers in Genesis, founder of a controversial Creation Museum in Kentucky.
The museum, which opened last year, depicts the Bible’s first book, Genesis, as literal truth. Its exhibits have been welcomed by those who believe that God created the heavens and the Earth in six days about 6,000 years ago.
Menton reckoned that Noah probably only had to take aboard about 16,000 creatures and said that most projected species discoveries are of tiny organisms.
“And we can leave out all organisms known to survive extensive flooding — such as insects and worms,” he said.
Even though creationists reject evolution, Menton said Noah may have taken along pairs to represent closely related “kinds” of animals such as dogs, wolves, coyotes and dingos, or just one pair for cows and buffaloes or tigers and lions.
He said Noah might have saved space by bringing along juveniles, including dinosaurs such as T-Rex or giant sauropods that could grow up to 30 metres (98 ft) long. Creationists believe that dinosaurs co-existed with humans.
The dimensions of the wooden Ark, given in cubits in the Book of Genesis, imply it was about 140 metres long, 23 wide and 13.5 high. It had three levels, meaning a total deck space of just short of 10,000 square metres (107,600 sq ft).
“The cargo capacity of such a ship would be impressive for those times and could be as large as 30,000 tons,” said Dragos Rauta, an expert at the International Association of Independent Tanker Owners (INTERTANKO).
Even so, the vessel would struggle to comply with modern marine transport guidelines, even with a few thousand creatures.
Noah and his family took along at least two of every type of animal and bird, and food for all on a voyage that lasted for months. The Bible also says that “clean” animals, or those deemed fit for eating such as cattle, sheep and goats, were taken in sevens.
Bjorn Clausen, managing director of Danish livestock shipping experts Corral Line AS, said large cows need at least 2 square metres each when held in pens for half a dozen animals.
“For animals like tigers, I’m not an expert but I’d say if you sail you’d need at least 4 square metres for a single tiger,” he said. What with other big animals such as kangaroos and rhinoceroses, the Ark would have quickly filled up.
HOT AND COLD
And zebras, penguins, vultures, pandas and antelope all need very different temperatures, food and habitats.
“Noah would have to be a very skilled heating and ventilation engineer … to have polar bears and iguanas on the same boat,” said Jesse Ausubel, chairman of the Encylopedia of Life at Rockefeller University in New York City.
“I’m not sure about the volume but … they wouldn’t all want the same conditions in their cabins,” he said.
The 2005 Millennium Ecosystem Assessment, a study by more than 1,300 scientists, estimated the number of identified species at 1.7-2 million with the final total likely to be between 5 and 30 million.
Of the named species, the biggest group by far, numbering around 1 million, are insects, centipedes and millipedes. Other big groups include plants, vertebrates — such as humans or whales — and molluscs. Mammals alone total more than 5,000 species — a few live in the sea, like whales.
In a line taken by creationists to argue that insects survived outside the Ark, Genesis says “everything on dry land that had the breath of life in its nostrils died” in the flood.
Insects do not have nostrils, so perhaps they survived by floating on uprooted trees or other debris.
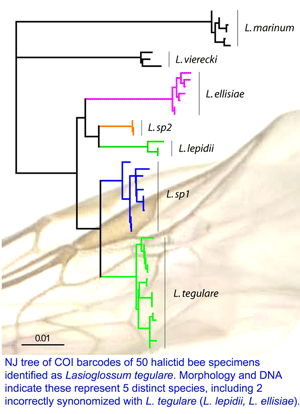 On 15 may 2008 an international assembly of bee experts gathered at York University and announced a new initiative to DNA barcode world’s bees. Some snippets from news reports:
On 15 may 2008 an international assembly of bee experts gathered at York University and announced a new initiative to DNA barcode world’s bees. Some snippets from news reports:
 Authors Garros, Ngugi, Githeko, Tuno, and Yan collected anopheline larva near Kisumu in western Kenya, dissected stomach contents of third and fourth instar forms, extracted DNA, and amplified an 800 bp fragment of nuclear 18s rRNA. A separate PCR assay was used to confirm species identity (five were A. gambiae s.s. and 68 were sister species A. arabiensis). According to authors, 18s rRNA was analyzed rather than COI because “more sequences are available [for 18s than for COI] in databases for plants, fungi, and protists”. I note there are now many research groups working on “plants, fungi, and protists” so it should be possible to achieve greater resolution in this sort of study as the DNA barcode libraries are built up.
Authors Garros, Ngugi, Githeko, Tuno, and Yan collected anopheline larva near Kisumu in western Kenya, dissected stomach contents of third and fourth instar forms, extracted DNA, and amplified an 800 bp fragment of nuclear 18s rRNA. A separate PCR assay was used to confirm species identity (five were A. gambiae s.s. and 68 were sister species A. arabiensis). According to authors, 18s rRNA was analyzed rather than COI because “more sequences are available [for 18s than for COI] in databases for plants, fungi, and protists”. I note there are now many research groups working on “plants, fungi, and protists” so it should be possible to achieve greater resolution in this sort of study as the DNA barcode libraries are built up.
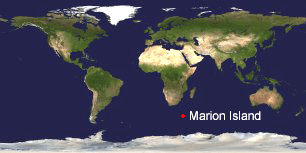 The challenge is to recognize invasive species before they become established. In
The challenge is to recognize invasive species before they become established. In  In April 2004, 3 noctuid moth larvae were found in an abandoned Wandering Albatross nest, a common habitat for one of the indigenous moth species. The larvae could be tentatively identified only to genus level and so rearing was attempted, with one larva dying after several months of pupating (as an aside, this is one example of how morphologic identifications can be laborious and/or incomplete, even for experts). The final larva was killed and preserved for DNA study; COI DNA barcode region was amplified using standard Folmer primers. The Marion Island moth larva barcode clustered with the 40 or so Black Cutworm Agrotis ipsilon sequences in BOLD, and was distinct from COI sequences of the other 18 Agrotis species in BOLD. Agrotis ipsilon is a common pest that feeds on a wide variety of plants. The authors conclude that Agrotis ipsilon is an established alien species with the potential to disrupt local ecosystems and that “steps be taken to eradicate the species from Marion Island.”
In April 2004, 3 noctuid moth larvae were found in an abandoned Wandering Albatross nest, a common habitat for one of the indigenous moth species. The larvae could be tentatively identified only to genus level and so rearing was attempted, with one larva dying after several months of pupating (as an aside, this is one example of how morphologic identifications can be laborious and/or incomplete, even for experts). The final larva was killed and preserved for DNA study; COI DNA barcode region was amplified using standard Folmer primers. The Marion Island moth larva barcode clustered with the 40 or so Black Cutworm Agrotis ipsilon sequences in BOLD, and was distinct from COI sequences of the other 18 Agrotis species in BOLD. Agrotis ipsilon is a common pest that feeds on a wide variety of plants. The authors conclude that Agrotis ipsilon is an established alien species with the potential to disrupt local ecosystems and that “steps be taken to eradicate the species from Marion Island.” Just posted, a
Just posted, a