How does one collect tropical reef fish without leaving North America? In July 2009 PLos ONE researchers from University of Guelph report on genetic diversity in SE Asian tropical reef fish, collected without plane fares or permits. How did they do it? Steinke and colleagues analyzed “dead on arrival” marine fish imported into Canada for the ornamental pet trade from various locations in SE Asia. A total of 1631 specimens representing 391 named species were frozen, imaged on a flatbed scanner, and a muscle tissue sample was taken for COI analysis. This is remarkable on several counts. First, the large number of species–according to FAO report cited by Steinke, “some 800 marine fish species, representing about 5% of all marine taxa, are involved in this trade, with 70% of sales directed to North America,” and estimated revenue of $200-$300 million annually. Second, this study surveys genetic biodiversity in reef fishes, provides a practical method for identification, and at the same time provides insight into what is probably the major threat to their survival. I am reminded of near extinction of Common egrets in North America in the late 1800’s as a result of hunting for plumes in women’s hats. This led to a popular uprising among women of fashion, who pledged not to wear such clothing, organizing what were the first “Audubon Societies” and successfully petitioning for legislative change, saving egrets and many other birds. Nemo and other reef fish may need a similar campaign.
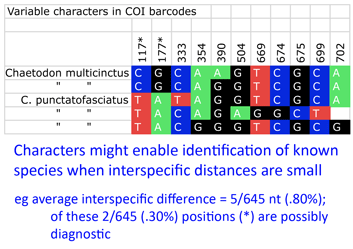 Back to the study, Steinke and colleagues found distinct barcodes among 384/391 (98.2%); 9 species displayed 2 or 3 distinct clusters, most of which were allopatric. Review of these potential “splits” revealed possible inappropriate synonymization in several cases. On the other side, 2 pairs and 1 triplet of species were not distinguished by DNA barcodes using distance. I look more closely at one of these examples, butterfly fishes Chaetodon multicinctus and C. punctatofasciatus, to see if there might be diagnostic characters whose signal is swamped by intraspecific variation. As in figure, there are 2 possibly diagnostic differences among this species pair. Of course, this sort of analysis only works for known species, but I wonder how many other species pairs/sets with “overlapping” barcodes have diagnostic differences.
Back to the study, Steinke and colleagues found distinct barcodes among 384/391 (98.2%); 9 species displayed 2 or 3 distinct clusters, most of which were allopatric. Review of these potential “splits” revealed possible inappropriate synonymization in several cases. On the other side, 2 pairs and 1 triplet of species were not distinguished by DNA barcodes using distance. I look more closely at one of these examples, butterfly fishes Chaetodon multicinctus and C. punctatofasciatus, to see if there might be diagnostic characters whose signal is swamped by intraspecific variation. As in figure, there are 2 possibly diagnostic differences among this species pair. Of course, this sort of analysis only works for known species, but I wonder how many other species pairs/sets with “overlapping” barcodes have diagnostic differences.
 In
In  In
In  In addition to economic and environment impact, mislabeling can have public health implications. In
In addition to economic and environment impact, mislabeling can have public health implications. In 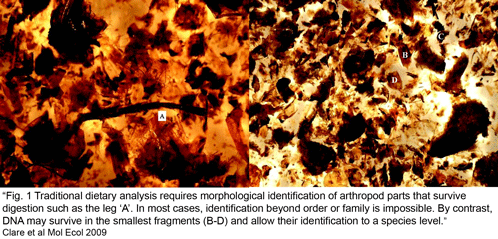 Clare et al obtained sequence data from 89% of 896 arthropod fragments; 78% of these were identified to species or genus level (the remaining 22% showed sequence similarity to bacteria, fungi, or were unidentifiable or chimeric), with a total of 127 prey species identified (125 insects, mainly lepidoptera including a number of economically important pest species, and 2 spiders). The “molecular scatology” approach documented greater diversity in prey species than prior studies based on morphologic analysis. Most prey were identified only once, with an average of 3.5 species per guano sample. Surprisingly, “more than 60% [of recovered insects] appear to have ears capable of hearing the echolocation hunting calls of L. borealis.” The authors speculate the abundance of eared moths might reflect bats hunting around streetlights, as moths in such brightly-lit environments are thought to use daytime predator-avoidance strategies rather than nocturnal responses to echolocation. There was a notable absence of actiid and tortricid moths, given their local abundance, suggesting these moths may have alternative predator-avoidance strategies.
Clare et al obtained sequence data from 89% of 896 arthropod fragments; 78% of these were identified to species or genus level (the remaining 22% showed sequence similarity to bacteria, fungi, or were unidentifiable or chimeric), with a total of 127 prey species identified (125 insects, mainly lepidoptera including a number of economically important pest species, and 2 spiders). The “molecular scatology” approach documented greater diversity in prey species than prior studies based on morphologic analysis. Most prey were identified only once, with an average of 3.5 species per guano sample. Surprisingly, “more than 60% [of recovered insects] appear to have ears capable of hearing the echolocation hunting calls of L. borealis.” The authors speculate the abundance of eared moths might reflect bats hunting around streetlights, as moths in such brightly-lit environments are thought to use daytime predator-avoidance strategies rather than nocturnal responses to echolocation. There was a notable absence of actiid and tortricid moths, given their local abundance, suggesting these moths may have alternative predator-avoidance strategies.