 The most successful technologies generate money. In turn, a commercial market helps drive improvements in cost and speed, enabling wider applications and new scientific knowledge. The rapid completion of the Human Genome Project (HGP) can be seen as a direct result of Applied Biosystems ABI 3700 DNA analyzer, the first fully automated capillary sequencer, introduced in 1998. In turn, the large market for high-throughput sequencing that resulted from HGP funding helped drive multiple rounds of improvement in cost and speed.
The most successful technologies generate money. In turn, a commercial market helps drive improvements in cost and speed, enabling wider applications and new scientific knowledge. The rapid completion of the Human Genome Project (HGP) can be seen as a direct result of Applied Biosystems ABI 3700 DNA analyzer, the first fully automated capillary sequencer, introduced in 1998. In turn, the large market for high-throughput sequencing that resulted from HGP funding helped drive multiple rounds of improvement in cost and speed.
This leads me to thoughts about DNA barcoding. The first exploratory meetings were held in 2003 at Banbury Center, Cold Spring Harbor Laboratory. Seven years later DNA barcoding is established as an accurate method for species identification with diverse scientific applications. BOLD, the publicly-available library of DNA barcodes, contains over 800,000 records from over 70,000 species. A new international effort, iBOL, is underway to establish DNA barcode libraries for 5 million specimens from 500,000 species by 2015. Like the government-maintained network of GPS satellites, publicly-funded DNA barcode libraries appear to offer enormous commercial opportunity, with potential benefits to society and science.
Where is barcoding on this path? So far, I find only a handful of companies and/or products that provide DNA-based species identification (for example, Therion, SteriSense, FishDNAID, Applied Food Technologies, Ecogenics). Of the few that exist, most are aimed at fish identification and do not take advantage of large scope and transparent sourcing of DNA barcode libraries. For example, Agilent Technologies recently introduced a “Fish identification system” based on “experimentally-derived [PCR-RFLP] patterns from more than 50 species.” This is wonderful but the scope is too small and the underlying library is unknown. Agilent is participating with the National Center for Food Safety and Technology, a US government-industry collaboration, so perhaps that will lead to more robust applications. I note that DNA barcode detection of food fraud (not just fish) was front-page news in Washington Post in March 2010 and the potential educational market is also large. I look forward to more entrepreneurs, whether at established companies or start-ups!
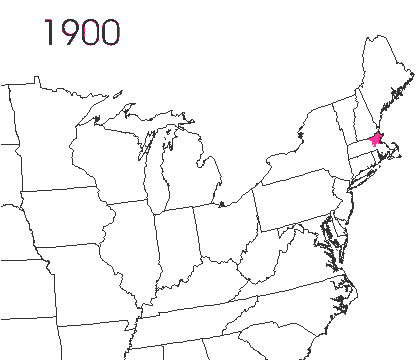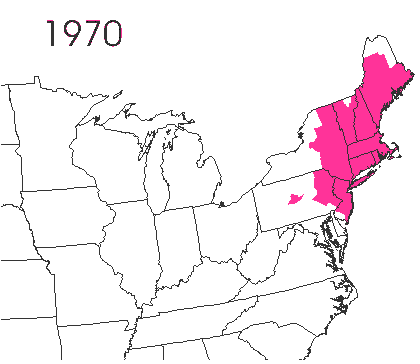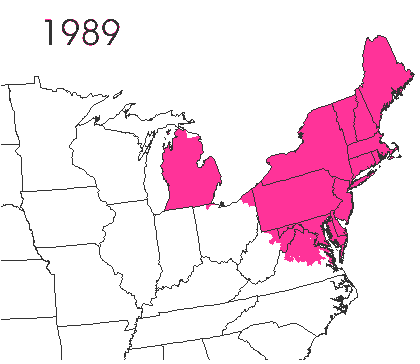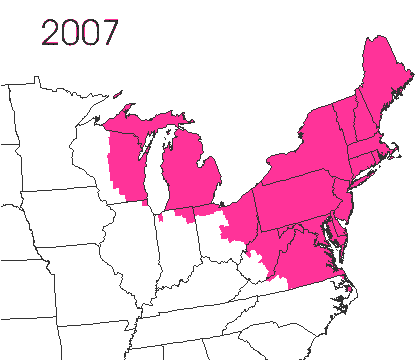 In the late 1860’s, a French entomologist, Étienne Léopold Trouvelot, living in Medford, Massachusetts, imported gypsy moths (
In the late 1860’s, a French entomologist, Étienne Léopold Trouvelot, living in Medford, Massachusetts, imported gypsy moths ( The first step in controlling invasive species is detection. In
The first step in controlling invasive species is detection. In 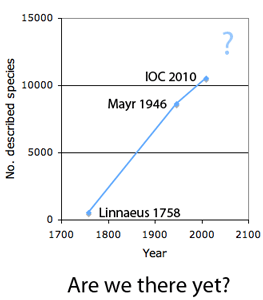 How many birds in the world? In the tenth edition of Systema Naturae (1758) (copy in US Library of Congress can be viewed or downloaded
How many birds in the world? In the tenth edition of Systema Naturae (1758) (copy in US Library of Congress can be viewed or downloaded 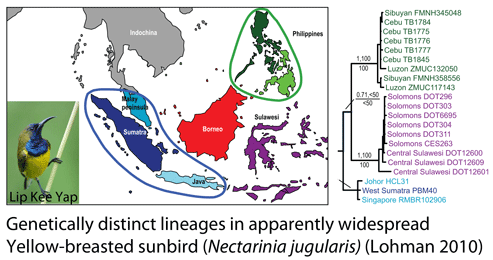 In some regions and categories of birds, the proportion of unrecognized species may be even higher. In
In some regions and categories of birds, the proportion of unrecognized species may be even higher. In 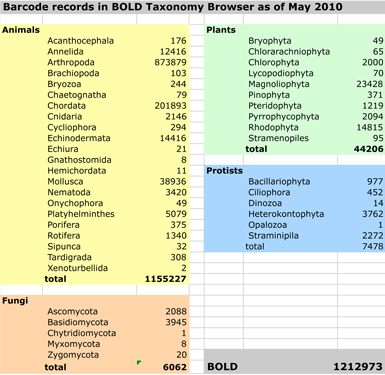 work with. Nonetheless, DNA barcoding of plants is ready for practical application and is providing immediately useful information (e.g. “DNA barcoding exposes a case of mistaken identity in the fern horticultural trade”
work with. Nonetheless, DNA barcoding of plants is ready for practical application and is providing immediately useful information (e.g. “DNA barcoding exposes a case of mistaken identity in the fern horticultural trade” 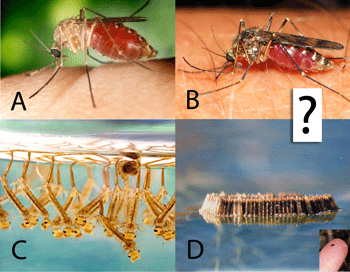 Biting insects transmit human and animal diseases, including protozoan (e.g., malaria, leishmania, trypanosoma (sleeping sickness, Chagas disease)), filiarial (e.g., onchocerciasis, Guinea worm), and viral (e.g., yellow fever, West Nile, dengue) diseases. Control measures rely on identifying the insects, which generally requires expert training.
Biting insects transmit human and animal diseases, including protozoan (e.g., malaria, leishmania, trypanosoma (sleeping sickness, Chagas disease)), filiarial (e.g., onchocerciasis, Guinea worm), and viral (e.g., yellow fever, West Nile, dengue) diseases. Control measures rely on identifying the insects, which generally requires expert training.