In October 2012 Nature 490:535, Breen and colleagues reported on amino acid variation among 13 mitochondrial protein and 2 nuclear proteins based on alignments of 3,000-53,000 sequences representing 1,000 to 14,000 species. They found that on average, a given site in a protein accomodates 9 different amino acids. Based on the distribution of variants, they conclude that epistasis (interaction among genes) strongly constrains molecular evolution.
Here Kevin Kerr and I re-analyze their large COI dataset [19,000 sequences (8,300 human); 4,700 species], generously provided by senior author Fyodor Kondrashov. Our aim is to determine if the frequency matrix approach we applied to avian BARCODEs (PLoS ONE 2012 e:43992) can be used to identify errors in a more phylogenetically diverse dataset. As the authors note, sequencing error is a potential confounder for their analysis; they used a different approach to assess error than we present here.
Brief methods. COI nucleotide alignment opened in MEGA, translated using appropriate table (~95% of COI dataset is insects or vertebrates), and exported to Excel; frequencies calculated at each amino acid position, and amino acid letter sequences converted into amino acid frequencies. For this analysis we defined rare variants as amino acids present in fewer than 0.02% (1/5000) sequences. In this dataset, rare variants comprised about half (46%) of the total amino acid diversity. For analyses illustrated below, we excluded the 8,281 human sequences, which had very few (8) rare variants.
Results
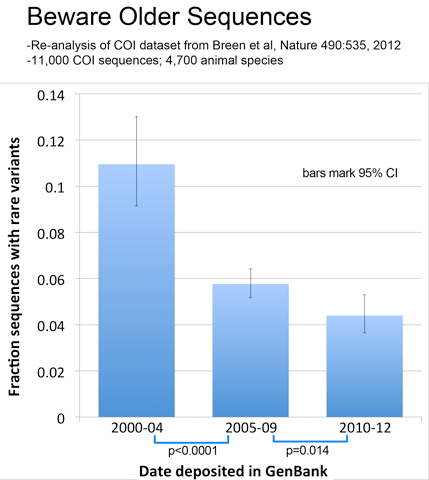
As observed with avian BARCODEs, rare variants in this dataset were less common in newer sequences, consistent with improved sequence quality over time.
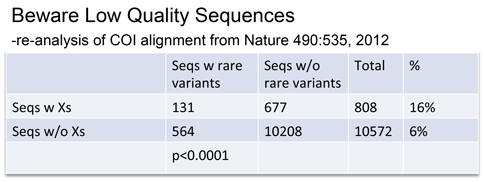
Rare variants were associated with low quality sequences–those with internal N’s, generating unknown “X” amino acids.
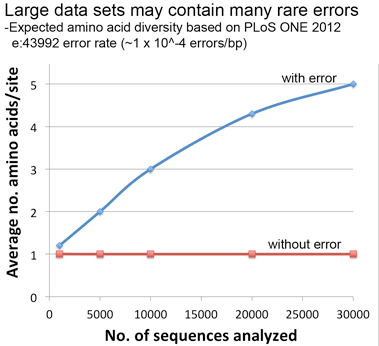
Lastly, a thought experiment applying the error rate from our PLoS ONE paper suggests that significant artifactual amino acid diversity is expected when error rate x dataset size is equal to or greater than 1, conditions that may be met by large datasets particularly those containing older sequences as in this COI alignment.
These results reinforce our published observation that a frequency matrix approach is a useful and important tool for analyzing error among large datasets. We hope that others will utilize this approach.
Regarding the findings of Breen and colleagues, our re-analysis suggests that error makes a greater contribution to amino acid diversity in this dataset than that calculated by authors, although the main conclusion of their paper regarding epistasis would likely be unchanged.