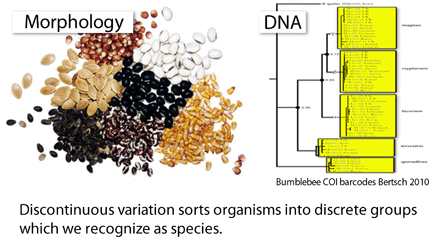
 Species are the units of biodiversity. Discontinuities in biological variation sort organisms into discrete groups that we recognize as species, and so gathering data on differences among organisms is the necessary first step in understanding the diversity of life. Here DNA has singular value–all organisms have DNA and some genetic loci are widely-shared, enabling direct comparisons across the diversity of multicellular life. Barcoding targets widely-shared gene sequence(s) that nonetheless differ among most closely-related species (COI for animals and rbcL+matK for land plants), providing broadly applicable metrics for mapping the discontinuities that represent species. Large-scale DNA barcoding thus offers for the first time a macroscopic view of biodiversity.
Species are the units of biodiversity. Discontinuities in biological variation sort organisms into discrete groups that we recognize as species, and so gathering data on differences among organisms is the necessary first step in understanding the diversity of life. Here DNA has singular value–all organisms have DNA and some genetic loci are widely-shared, enabling direct comparisons across the diversity of multicellular life. Barcoding targets widely-shared gene sequence(s) that nonetheless differ among most closely-related species (COI for animals and rbcL+matK for land plants), providing broadly applicable metrics for mapping the discontinuities that represent species. Large-scale DNA barcoding thus offers for the first time a macroscopic view of biodiversity.
This sounds straightforward enough, but naming species, like medical diagnosis, is a process requiring human judgment. A taxonomic expert generally focuses on one or a few species or potential species at a time, sifting through morphological, ecological, behavioral, and DNA data and making inferences about the evolutionary past. It generally takes years or decades between specimen collection and publication of a new species description, and my impression is that most specimens in museum collections including frozen tissues have never been scrutinized in detail sufficient to determine whether they represent new species. Given that a high-throughput laboratory can generate a hundred thousand barcodes in a year, there are opportunities for new workflows.
In May 2010 Frontiers Zool, researchers from Uppsala University, Sweden, and Technical University of Braunschweig, Germany, look at how we might incorporate the flood of DNA data, outlining an approach they (and others) call “integrative taxonomy”. As current practice in taxonomy already involves integrating different kinds of data (morphology, behavior, range, DNA), I take this term to mean an approach somewhere between one primarily based on morphology (“traditional taxonomy”) and one primarily based on DNA (“DNA taxonomy”), such as that for eubacteria and archaebacteria. Padial and colleagues review the recently revitalized scientific discussion about species delimitation involving population biology and phylogenetics, noting “what matters for the study of speciation matters for taxonomy as well.” They call for a flexible approach including the possibility of “recognition of a species on the basis of a single set of characters”, which could be DNA barcodes. Near the end, they address the big challenge, which is that DNA studies, particularly DNA barcoding, “are revealing units that might represent potential new species at a faster pace than results can be followed up for taxonomists.” Padial and co-authors review various protocols used for naming “candidate species” and conclude “standardization of such schemes across taxonomic groups of eukaryotes would be clear progress for data retrieval systems.” As described in more detail here previously, a starting point for discussion of the preferred format for standardizing provisional names was recently proposed (Schindel and Miller, System Naturae 250, Chapter 10), based on the scheme currently used by CHAH (Council of Heads of Australian Hebaria). This system of “taxon labels” (as distinguished from “taxon names”) meets the criteria of uniqueness, stability, and non-confusion with formal taxon names.
At present, our knowledge of biodiversity is built around a catalog of taxon names, annotated with DNA data if available. I imagine the future catalog as being a DNA (barcode) map, annotated with taxon names if available. Some parts of the map, such as for birds, will be heavily annotated, and others, such as for nematodes, will have few formal names, and instead will have taxon labels generated by automated clustering algorithms. In some cases, the DNA data will be derived from individual specimens, backed up by museum vouchers, and in other cases it will be generated from environmental sampling. Only then will we begin to see how much biodiversity is unexplored.