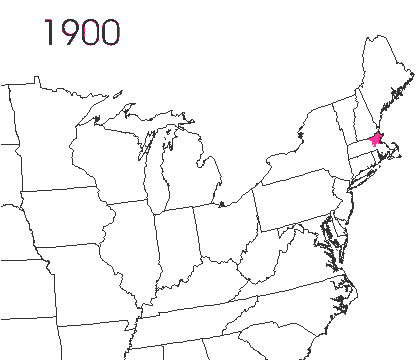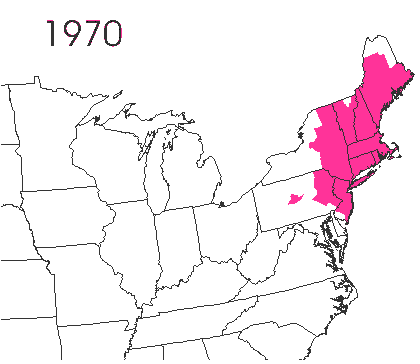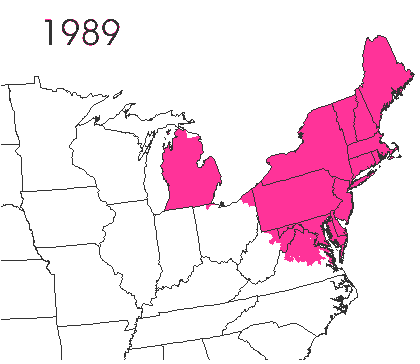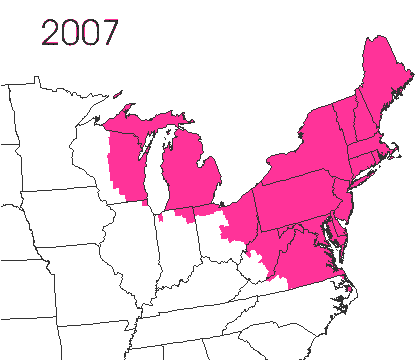
 In the late 1860’s, a French entomologist, Étienne Léopold Trouvelot, living in Medford, Massachusetts, imported gypsy moths (Lymantria dispar) which he hoped to hybridize with domesticated Asian silkworms (Bombyx mori), thereby creating a new silk-producing strain with improved disease resistance (for history, see US Forest Service page). The experiment failed (not surprising given moths are from different families), the colony escaped from Trouvelot’s backyard, and gypsy moths became established as a major pest of hardwoods in the northeastern US (animated range data from US Forest Service at right). Subsequent introductions of numerous forest pests and pathogens into the US, largely through importation of infested wood products, have had large impacts on timber industry and local ecosystems alike, and have led to near extinction of American chestnut, and large-scale mortality in elm, hemlock, and oak, and other tree species.
In the late 1860’s, a French entomologist, Étienne Léopold Trouvelot, living in Medford, Massachusetts, imported gypsy moths (Lymantria dispar) which he hoped to hybridize with domesticated Asian silkworms (Bombyx mori), thereby creating a new silk-producing strain with improved disease resistance (for history, see US Forest Service page). The experiment failed (not surprising given moths are from different families), the colony escaped from Trouvelot’s backyard, and gypsy moths became established as a major pest of hardwoods in the northeastern US (animated range data from US Forest Service at right). Subsequent introductions of numerous forest pests and pathogens into the US, largely through importation of infested wood products, have had large impacts on timber industry and local ecosystems alike, and have led to near extinction of American chestnut, and large-scale mortality in elm, hemlock, and oak, and other tree species.
 The first step in controlling invasive species is detection. In J Entomolog 2010 7:60 researchers from USDA Forest Service report on DNA barcode identification of Eurasian woodwasp Sirex noctilio. S. noctilio has been established and spreading in northeastern US and Canada since at least 2004, and “will likely become a major pest of pines and possibly other conifers in North America.” The wasp attacks living pines, laying eggs along with an inoculum of “phytotoxic mucus” and an exotic [non-native] wood decay fungus (Amylosterum areolatum). The wasp larvae “feed on pine wood decayed by the fungus and on the fungus itself”, weakening or killing the tree.
The first step in controlling invasive species is detection. In J Entomolog 2010 7:60 researchers from USDA Forest Service report on DNA barcode identification of Eurasian woodwasp Sirex noctilio. S. noctilio has been established and spreading in northeastern US and Canada since at least 2004, and “will likely become a major pest of pines and possibly other conifers in North America.” The wasp attacks living pines, laying eggs along with an inoculum of “phytotoxic mucus” and an exotic [non-native] wood decay fungus (Amylosterum areolatum). The wasp larvae “feed on pine wood decayed by the fungus and on the fungus itself”, weakening or killing the tree.
Wilson and Schiff analyzed COI barcodes of 207 larvae or adults representing 27 woodwasp species or subspecies (including 6 Sirex spp.) following a fairly standard protocol (i.e., 1 leg, DNAeasy kit, HCO 2198/LCO 1490 primers.) [As an aside, these primers (Folmer 1994) remain surprisingly widely used for barcoding invertebrates, despite development of several other effective broad-range primers for COI barcode region (e.g., see CCDB collected protocols), which perhaps reflects absence of a large-scale direct comparison.] All species gave distinct barcodes, minimum interspecific distance was 7.6 (maximum 26.2%) , and, remarkably, there was no variation within any named taxa (average 9 individuals per species/subspecies, range 4-23). However they observed 2.3%-2.8% differences between subspecies of Xeris spectrum and Sirex juvencus, suggesting that “taxonomic revisions are probably in order to separate these subspecies in each case into separate subspecies.”
In addition to application in forest surveys, Wilson and Schiff note the need for a “standardized diagnostic method of identifying insect larval stages at ports of entry within imported wood producs…and in wood used as crates and dunnage for imported goods.” For example, “recent analyses of Sirex larvae intercepted from 1985-2000 by USDA-APHIS personnel at US ports of entry…indicate that only 7 (6.8%) of 103 specimens could be identified to species (Hoebeke et al 2005).” The authors conclude “DNA barcode methods can be used to identify larval states of woodwasps…as easily as free-flying adults,” which “should help prevent future introductions of S. noctilio and other exotic woodwasps.”