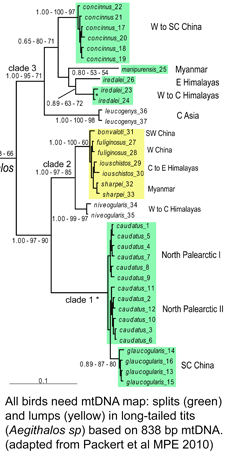
 Although birds have been studied in more detail than any other large group of animals, mtDNA continues to reveal many overlooked species, such that named taxa turn out to be comprised of two or more distinct species. These revisions include some very familiar birds, e.g., Canada Goose, which was recently recognized as comprising two species, Cackling Goose (B. hutchinsii) and Canada Goose (B. canadensis) (A.O.U. Check-list 45th suppl. 2004); for a current example, see Päckert et al Mol Phylogenet Evol 2010). Although such taxonomic revisions reflect a combined analysis of morphological, behavioral (particularly song), geographic range, and DNA information, to my reading mtDNA generally trumps the other data, which mostly serve as corroborating evidence. It is not that mtDNA similarities or differences are important per se, it is that they are strongly predictive of the presence or absence of organismal differences, particularly reproductive isolation, that are the hallmarks of species status. Of the species examined so far (I estimate about 1/3 of the 10,000 world birds), most demonstrate a similar patterning of limited mtDNA differences within species and relatively large differences among species, so we can be confident that this analytic approach will hold up. As an aside, referring to splits of named species as “taxonomic inflation” is misleading, as it suggests the real number of species is already known, which seems no more correct than referring to binary stars resolved with a new telescope as reflecting “stellar inflation.”
Although birds have been studied in more detail than any other large group of animals, mtDNA continues to reveal many overlooked species, such that named taxa turn out to be comprised of two or more distinct species. These revisions include some very familiar birds, e.g., Canada Goose, which was recently recognized as comprising two species, Cackling Goose (B. hutchinsii) and Canada Goose (B. canadensis) (A.O.U. Check-list 45th suppl. 2004); for a current example, see Päckert et al Mol Phylogenet Evol 2010). Although such taxonomic revisions reflect a combined analysis of morphological, behavioral (particularly song), geographic range, and DNA information, to my reading mtDNA generally trumps the other data, which mostly serve as corroborating evidence. It is not that mtDNA similarities or differences are important per se, it is that they are strongly predictive of the presence or absence of organismal differences, particularly reproductive isolation, that are the hallmarks of species status. Of the species examined so far (I estimate about 1/3 of the 10,000 world birds), most demonstrate a similar patterning of limited mtDNA differences within species and relatively large differences among species, so we can be confident that this analytic approach will hold up. As an aside, referring to splits of named species as “taxonomic inflation” is misleading, as it suggests the real number of species is already known, which seems no more correct than referring to binary stars resolved with a new telescope as reflecting “stellar inflation.”
As with most animal groups, there are no nuclear genes that regularly distinguish closely-related birds. (It seems likely that sequencing entire nuclear genomes will enable discriminating species units, but this is inherently a more costly, less standardizable approach, making it unattractive for routine use.) Differences among closely-related species are more or less evenly-distributed throughout the mitochondrial genome, so that approximately 500-1000 bp of coding DNA usually contains sufficient information to recognize species and create genus-level phylogenies. Moreover, if species are not distinguished by this length of mtDNA then additional sequencing usually does not improve the resolution. This might be better stated the other way around, namely, if two sets of birds cannot be distinguished by a relatively short stretch of mitochondrial DNA, then they most likely belong to the same species. Why is this so? The prevailing null hypothesis is that mitochondrial differences are an accidental if useful accompaniment to species status, reflecting genetic drift over the several hundred thousand years of reproductive isolation presumably required for new species to emerge. In a recent essay (Nature 18 nov 2009), Nick Lane explores the interesting possibility that mitochondrial differences cause speciation, thus producing what they identify.
What barcoding adds to the historical approaches in avian systematics is standardization, so that the same locus is analyzed in every specimen, which speeds completion of species-level avian taxonomy and facilitates large-scale comparisons. Given that most bird species have yet to analyzed for species-level genetic differences, the standardized approach can save considerable time and money, as the results of independent investigators can be readily merged (e.g. Johnsen et al J Ornithol 2010, Kerr et al Frontiers Zool 2009). Beyond classification, this approach creates a reference library of avian barcodes, which enables identifying unknown specimens, such as from birdstrikes (Marra et al Frontiers Ecol Environ 2009). Our survey of avian frozen tissue collections identified over 315,000 specimens representing over 7,200 species (Stoeckle and Winker, Auk 2009). Based on what is in GenBank, it appears that most of these specimens have never been analyzed for any genetic locus, so a first-pass effort to sequence COI barcode region will certainly reveal many new species and help resolve higher-level relationships as well. There is an opportunity for a granting agency or foundation to have a large impact on avian systematics for modest cost by supporting barcoding of existing collections, given that the tissue specimens and vouchers, which are the most expensive components of collections, have already been prepared. Looking further ahead, such an effort would also create sets of high-quality DNA extracts, with species identity confirmed by COI barcode, linked to vouchered specimens, that would be a powerful resource for further genetic study.