The entire May 2009 Mol Ecol Res “Special Issue on Barcoding Life” is open access, thanks to support from Genome Canada and NSERC. As an aside, Mol Ecol Res publisher Wiley-Blackwell, which puts out over 1400 journals, charges $3000 US per article for open access, as compared to, for example, $1300 in PLoS ONE (all articles open access), and $1200 (plus $70/page) for open access option in Proc Natl Acad Sci USA. If funders mandate open access for publications based on research they support, then either this differential will disappear, or many manuscripts will migrate to lower cost journals. The special barcoding issue is based on Canadian Barcode of Life Network Scientific Symposium held at the Royal Ontario Museum in April 2008 and includes 27 articles on topics ranging from methodology to applications in creatures great and small including fungi and plants.
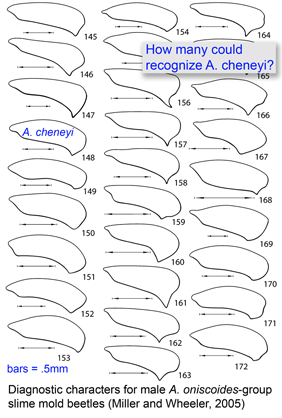 Most DNA barcoding analyses look at DNA identification through the lens of established taxonomy, ie how well does sequence data capture the species-level taxonomic categories established by morphologic analysis? In the special issue article “DNA barcoding and the mediocrity of morphology” researchers from York University and University of Guelph look at the comparison the other way around–how well does morphology identify the sorts of specimens that can be distinguished by DNA-based methods, barcoding in particular? In Packer and colleagues’ analysis, morphology comes up short “in numerous important situtations such as the association of larvae with adults and discrimination among cryptic species.” Taking an example not entirely at random, the authors analyze a key to Agathidium genus slime mold beetles co-authored by a sometime skeptic of barcoding (Miller and Wheeler, 2005) (this key made popular news as 3 of newly described beetles were named in tribute to then current US government officials–A. bushi, A. cheneyi, A. rumsfeldi). As is common in keys to insect identification, the reliance on adult male characters, usually genitalia, means that females and immature forms often cannot be identified to species (for the 3 USG namesakes, the key states “female not examined” and there is no description of immature forms). Again typical of insect keys, there is no documentation of intraspecific variation in diagnostic characters (for A. cheneyi, “the holotype is the only specimen examined of this species”). As a result, Packer and colleagues note “the morphological equivalent of the barcode gap that enables molecular identification of species cannot be calculated using traditional approaches, and the sample size of illustrations upon which measures of intraspecific variation might be estimated usually averages one per species with zero variance.”
Most DNA barcoding analyses look at DNA identification through the lens of established taxonomy, ie how well does sequence data capture the species-level taxonomic categories established by morphologic analysis? In the special issue article “DNA barcoding and the mediocrity of morphology” researchers from York University and University of Guelph look at the comparison the other way around–how well does morphology identify the sorts of specimens that can be distinguished by DNA-based methods, barcoding in particular? In Packer and colleagues’ analysis, morphology comes up short “in numerous important situtations such as the association of larvae with adults and discrimination among cryptic species.” Taking an example not entirely at random, the authors analyze a key to Agathidium genus slime mold beetles co-authored by a sometime skeptic of barcoding (Miller and Wheeler, 2005) (this key made popular news as 3 of newly described beetles were named in tribute to then current US government officials–A. bushi, A. cheneyi, A. rumsfeldi). As is common in keys to insect identification, the reliance on adult male characters, usually genitalia, means that females and immature forms often cannot be identified to species (for the 3 USG namesakes, the key states “female not examined” and there is no description of immature forms). Again typical of insect keys, there is no documentation of intraspecific variation in diagnostic characters (for A. cheneyi, “the holotype is the only specimen examined of this species”). As a result, Packer and colleagues note “the morphological equivalent of the barcode gap that enables molecular identification of species cannot be calculated using traditional approaches, and the sample size of illustrations upon which measures of intraspecific variation might be estimated usually averages one per species with zero variance.”
I hope that future keys for slime mold beetles will include DNA barcode sequences. This will enable anyone, scientists and public alike, with access to a DNA sequencer to identify A. cheneyi adults of both sexes, larvae, fragments in the guts of predators, and perhaps eggs in random leaf litter samples.
For a biology hobbyist, this site is a fascinating read. Through DNA barcoding, could we be taking huge steps towards effective cloning?