 Deep space telescopes gather light from the early universe, providing pictures of the unimaginably remote past. What about the biological universe–can we peer back in time? Geochemical evidence suggests life on Earth arose about 3.5 billion years ago and fossils reveal what life looked like as far back as 3.0 billion years, and important fossil discoveries across that whole span of time continue to be made. What about DNA? As Carl Woese first realized, DNA sequences of living organisms contain signatures of their evolutionary relationships, and enable reconstructing history as far back as the origin of replication, even before cells and DNA. At the near end of the time scale, recovery of DNA from historical samples can help identify organisms that lived hundreds, thousands, tens of thousands, or even, in a few cases so far, hundreds of thousands years ago.
Deep space telescopes gather light from the early universe, providing pictures of the unimaginably remote past. What about the biological universe–can we peer back in time? Geochemical evidence suggests life on Earth arose about 3.5 billion years ago and fossils reveal what life looked like as far back as 3.0 billion years, and important fossil discoveries across that whole span of time continue to be made. What about DNA? As Carl Woese first realized, DNA sequences of living organisms contain signatures of their evolutionary relationships, and enable reconstructing history as far back as the origin of replication, even before cells and DNA. At the near end of the time scale, recovery of DNA from historical samples can help identify organisms that lived hundreds, thousands, tens of thousands, or even, in a few cases so far, hundreds of thousands years ago.
In April 2009 PLoS ONE ten researchers from university centers in Denmark, United Kingdom, United States, Canada, Russia, and New Zealand report on non-destructive recovery of diagnostic DNA from ancient insect specimens. As an aside, PLoS ONE is an important sea change in scientific publishing. First of all, as described on their website, the journal “features reports of original research from all disciplines within science and medicine. By not excluding papers on the basis of subject area, PLoS ONE facilitates the discovery of the connections between papers whether within or between disciplines.” Second, it puts the judgement of importance in the hands the scientific community where it belongs:
 “Too often a journal’s decision to publish a paper is dominated by what the Editor/s think is interesting and will gain greater readership — both of which are subjective judgments and lead to decisions which are frustrating and delay the publication of your work. PLoS ONE will rigorously peer-review your submissions and publish all papers that are judged to be technically sound. Judgments about the importance of any particular paper are then made after publication by the readership (who are the most qualified to determine what is of interest to them).”
“Too often a journal’s decision to publish a paper is dominated by what the Editor/s think is interesting and will gain greater readership — both of which are subjective judgments and lead to decisions which are frustrating and delay the publication of your work. PLoS ONE will rigorously peer-review your submissions and publish all papers that are judged to be technically sound. Judgments about the importance of any particular paper are then made after publication by the readership (who are the most qualified to determine what is of interest to them).”
This is so sensible it is surprising it has not happened earlier! There is of course a place for journals like Nature and Science, but I expect that a great deal of scientific publishing will migrate to PLoS ONE, with benefits to the authors and the scientific community.
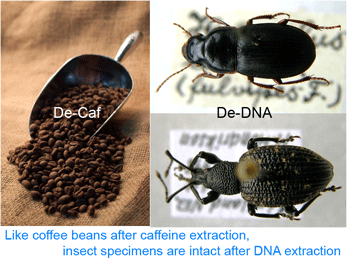
Back to the paper. Thomsen and colleagues first tested a non-destructive extraction method (Gilbert et al 2007 PLoS ONE 2:e272) on museum beetle specimens. This involves overnight incubation with gentle agitation in a digestion buffer at 55^o C. Remarkably, the specimens emerged none the worse for the wear. The researchers recovered 77-204 bp segments of mtCOI from all of 20 beetles, which were collected as early as 1825 (1/3 were over 100 years old). Using a Bayesian approach that generates taxonomic assignments with probability estimates, these short fragments were sufficient for identification to species in most cases; the remainder could be assigned to family or genus level. The researchers then applied this same technique to insect chitin (exoskeleton) fragments preserved in permafrost dating from about 7,000 to over 47,000 years before present (BP). Here only 3 of the 14 (21%) samples (10,000-26,000 y BP) yielded amplifiable DNA, with Bayesian assignments to family or order level. Although the authors appear to have hoped for higher success, this seems pretty remarkable to me. They speculate that destructive sampling might have produced higher yields.
Saving what might be the best for last, Thomsen and colleagues tested non-frozen sediment samples that lacked visible insect parts collected in New Zealand caves and dated 1800 to 3280 years BP. Using a more or less standard extraction protocol developed by some of the authors (Willerslev et al 2003 Science 300:791), 96 bp fragments of COI (1 beetle, 1 butterfly) were recovered from 2 of 3 samples tested. The authors drily note “although the non-frozen sediment DNA approach involves destructive sampling, it has the advantage that the material is the sediment itself, which is usually abundant, and normally not too valuable to process.”
I conclude that if bits of DNA are preserved in ancient dirt then DNA from the past and present must be all around us. Perhaps single molecule sequencing methods will reveal an even greater abundance and diversity of DNA in environmental samples.
Mehrdad Hadjibabei points out primers used in this study amplify segments of 3′ portion of COI. For future work, I hope the authors and others taking a similar approach utilize primers that amplify 5′ COI. This will allow them to take advantage of what is the largest reference database for any mtDNA region, ie DNA barcodes based on 5′ COI (>50K species, >500,000 individuals so far).