In 1 Oct 2008 Syst Entomol researchers from University of Alberta report on “Widespread decoupling of mtDNA variation and species integrity in Grammia tiger moths.” Authors Schmidt and Sperling analyzed COI sequences from 274 specimens representing 28 of 36 known Grammia species, collected across Canada and US. An NJ tree showed 13 haplogroups (loose clusters); 11 of these “largely or exclusively corresponded to nominal species,” while the other two, “designated the Western and Eastern haplogroups, contained polyphyletic asemblages of 13 and 10 species, respectively.” The researchers conclude that these two tangles of sequences and species represent historical or ongoing mating between species and “research on factors governing hybridization would be particularly informative in gaining an understanding of the role of isolating mechanisms in speciation” (ie DNA barcoding highlights an interesting group for further study).
Like explorers mapping new territory, Schmidt and Sperling’s study creates a map that can be used by the next investigators studying these moths, whether as eggs, larva (according to Caterpillars of Eastern North America by David Wagner, Princeton University Press, 2005 “there are no keys that can be used to identify the [Grammia] caterpillars with reliability”), intact adults, or as fragments retrieved from droppings of predator species such as bats. Specimens with COI barcodes in the two polyphyletic tangles will at least be identifiable to a subset of species within the genus.
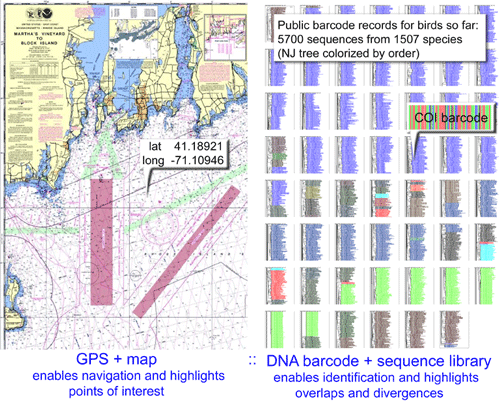
This study brings to mind an analogy between GPS and DNA barcodes. Handheld GPS devices enable us to pinpoint our location on the earth’s surface within a few meters. We then use a map to translate the numerical coordinates into useful information. In areas where the mapping is incomplete or out of date, GPS coordinates are less informative and may be misleading.
A DNA sequencer (handheld version soon perhaps) is a “biodiversity GPS” device, a DNA barcode is a set of biodiversity GPS coordinates, and a barcode reference library is a biodiversity map on which the specimen’s taxonomic identity can be located. In areas that have been mapped in detail (ie records from multiple specimens across the species range and from closely-related species), a barcode sequence will usually enable precise species-level identification with a high degree of certainty. In groups less-well surveyed or in which the taxonomy is unknown, there will be more uncertainty. Nonetheless, the general coherence of genera, families, and even orders in simple COI NJ trees (see figure below) suggests a DNA barcode will usually provide useful taxonomic information even in the absence of comprehensive taxonomic coverage.
I expect that in the future there will be good methods for defining species based on sequence data, including COI barcode records. While the importance of genetic data as an indicator of species status is informally recognized in science reporting (eg “DNA analysis confirmed it was a new species”), it is generally relegated to an ancillary role in species descriptions. It is remarkable to me that of all the mathematical tools of phylogeography, population genetics and phylogenetic reconstruction, none are designed to diagnose species. Just as a node in a ML tree may have 90% bootstrap support, why not apply the same rigor to species-level genetic data and say, for example, 90% confidence that this particular cluster represents a distinct species. I understand this would involve adopting a particular species concept, but at least it would be a place to start. If the data were only COI or other mtDNA sequences, then there might need be a warning about possible introgression as the above study demonstrates. I believe the flood of data from the barcode initiative, with multiple sequences from tens or hundreds of thousands of species, will help push development of such tools.
Google Earth, Google Sky, Google Ocean and why not a Google Species? It would really help a lot.
I would have to agree with Peanar. Google Species sounds quite nice.
@peanar and @sportsbook review. got news for you. google’s not going down that road. they can’t monetize. or even sponsor something like that.
@Charlotte SEO Guy. That is very true.. it would be a very expensive venture.
It would be an expensive venture, but I believe that google can monetize it. There are thousands of searches for this which would produce all kinds of advertisements that don’t even have to be relevant to the species.