By identifying species (leaves) and determining how they are related (branches), taxonomy aims to reconstruct the Tree of Life. To do so taxonomists must distinguish variation within species from that between species, and identify the shared characters that reflect evolutionary ancestry. These tasks require highly-specialized sets of knowledge and skill for each animal and plant group.
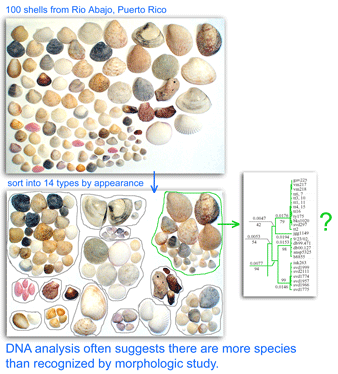 One result is that it has been difficult to compare patterns of diversification between different branches in the Tree. One might ask, how finely and how evenly divided is biodiversity? Are the differences among and within mosquito species (3,400 species), for example, similar to the differences among and within fruit flies (6,200 species) or birds (10,000 species)? Broad application of DNA analysis is beginning to provide some insights. To enable these sorts of comparisons, a standardized locus is needed, as unique genes can solve local branching patterns, but do not allow easy comparisons between branches.
One result is that it has been difficult to compare patterns of diversification between different branches in the Tree. One might ask, how finely and how evenly divided is biodiversity? Are the differences among and within mosquito species (3,400 species), for example, similar to the differences among and within fruit flies (6,200 species) or birds (10,000 species)? Broad application of DNA analysis is beginning to provide some insights. To enable these sorts of comparisons, a standardized locus is needed, as unique genes can solve local branching patterns, but do not allow easy comparisons between branches.
Large-scale surveys of standardized genetic loci, including COI barcoding, commonly reveal distinct groups within what was thought to be a single species. There is also the converse finding that in some cases COI barcode sequences do not distinguish named species, but generally this strikes me as a relatively minor scientific problem that usually involves very closely-related species pairs and can be solved where needed with more DNA sequence, assuming the underlying taxonomy is correct and the named species really are distinct. The greater scientific challenge is finding multiple groups within what appear morphologically to be single species. In many cases so far, organisms with genetically distinct COI barcode clusters show associated biological differences signalling they represent different species.
In December 2007 Mol Ecol 16:4999, researchers from Museum of Comparative Zoology, Harvard, examine genetic differences in Aoraki denticulata, a tiny (2-3 mm) daddy longlegs or harvestman spider found in leaf litter widely through New Zealand. They determined COI barcode region sequences for 119 individuals from 17 localities in the mountainous northern part of South Island. The two described subspecies A. d. denticulata and A. d. major were genetically distinct. The surprising finding was there were at least 14 distinct clusters within A. d. denticulata, with a different group at almost every site, and 2 clusters at 3 of the sites. The differences between mtDNA clusters were as larger or larger than between other Aoraki species, up to 19.2%, but no morphologic differences were found even with electron microscopic scanning of males. Boyer et al observe “while it is conceivable that some of the geographically widespread populations…represent cryptic species, it is difficult to imagine that morphologically identical individuals from a single sample at a unique geographical point are not conspecific…it is hard to believe that almost every sampled locality would host at least one, if not two, cryptic species.”
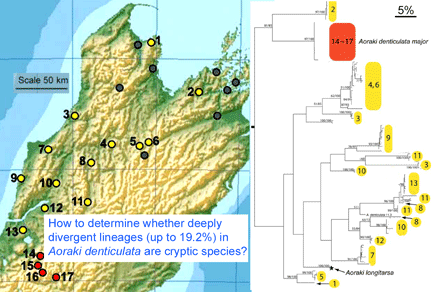
The authors helpfully provide scanning electron micrographs of the morphologic differences that do exist among the two subspecies of A. denticulata and other Aoraki species. To a non-specialist, these appear so minor that one can imagine there might be other more consequential differences in behavior or chemical signalling that are not visible but are associated with the observed genetic divergences. The authors suggest another alternative explanation, rapid sequence evolution, but this seems unlikely in that the differences within groups should also be larger, which does not appear to be the case. Whole genome sequencing is increasingly inexpensive and might enable determination of whether the A. denticulata clusters represent different species, and might be more practical than on-site observation of the behavior of very small creatures that inhabit leaf litter. I look forward to learning more about whether these spiders are exceptions to the rule that deep mtDNA divergences signal species boundaries in animals.