The movement to create a library of DNA barcodes for plants, animals, and fungi began with the Cold Spring Harbor Banbury meeting that Jesse Ausubel and Mark Stoeckle helped organize in 2003. During the 17-21 September 2007 the Consortium for the Barcode of Life convened its 2nd International Conference in Taipei along with meetings of working groups concerned with fish, all forms of marine life, plants, fungi, regional initiatives, and techniques. The progress, reported in a press release and covered in The Economist and also recounted in Mark’s Blog, is thrilling. The happy mood of the exciting conference shows in the photo of attendees. Thanks to Kwang-Tsao Shao (Academia Sinica), David Schindel (Consortium for the Barcode of Life), Karen Armstrong (New Zealand, chair of conference program committee), and many others for making a great success.
Blog
Taxonomy without borders
 341 researchers from 44 countries gathered for the Second International Barcode of Life Conference, held at Academia Sinica, Tapei, Taiwan on 17-21 September 2007 (program, participants, and abstracts at www.dnabarcodes2007.org).
341 researchers from 44 countries gathered for the Second International Barcode of Life Conference, held at Academia Sinica, Tapei, Taiwan on 17-21 September 2007 (program, participants, and abstracts at www.dnabarcodes2007.org).
Conference presentations highlighted a thrilling array of progress on diverse scientific and practical fronts since the First International Barcode of Life at The Natural History Museum, London, in February 2005 (London Conference proceedings in themed issue Phil Trans R Soc 360: 2005 available through Consortium for Barcode of Life (CBOL) website. I found the Tapei conference to be a landmark demonstration of the value to society and science of a standardized, inexpensive approach to identifying species through DNA, ie DNA barcoding. The Economist’s 20 September 2007 piece “Name, rank, and serial number” recaps results so far and looks ahead to near future societal benefits.
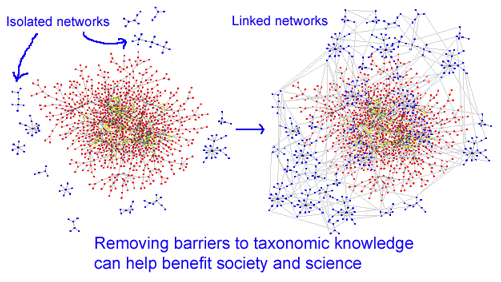
Near the close of the conference, David Schindel, Executive Secretary for CBOL, referred to the DNA barcode initiative as “taxonomy without borders”. Just as removing security fences benefits African wildlife, standardized inexpensive technology for species identification, ie DNA barcoding, is helping remove barriers that balkanize taxonomy and limit public access to biological knowlege. The DNA barcode initiative, together with the Encyclopedia of Life which includes digitizing the world’s taxonomic literature are creating powerful new ways of seeing biodiversity, with benefits to society and science.
I look forward to a future in which the multiple sectors of taxonomic and biodiversity science are densely linked to each other and public users.
Werewolf Microbes
“Werewolf Microbes might take power!” The e-zine Energy Tribune interviewed Jesse for its subscribers. We post the entire interview conducted by Robert Bryce.
Exploring mitochondrial DNA differences within species
Paul De Barro, CSIRO, Australia, recently posed the question “What is the expected level of mitochondrial variation within species?” The answer may be “almost none”. Results so far with DNA barcoding initiatives show average intraspecific variation in most animal species, whether saturniid moths or sand martins, is on the order of 0.5% or less. Here is my somewhat speculative set of inferences drawn from the finding of low variation within most animal species:
 1. Low intraspecific variation implies low effective population size (Ne); according to my back-of-the-envelope math, about 10,000 or so for most animal species. The apparent ceiling on Ne is low enough that census population size and species age, both of which might be expected to be determinants, do not contribute to intraspecific variation.
1. Low intraspecific variation implies low effective population size (Ne); according to my back-of-the-envelope math, about 10,000 or so for most animal species. The apparent ceiling on Ne is low enough that census population size and species age, both of which might be expected to be determinants, do not contribute to intraspecific variation.
2. What about species with larger average differences in mitochondrial DNA? Most are mosaics of reproductively-isolated or partially reproductively-isolated populations, some of which might be considered separate species. According to standard models of sequence evolution, it takes tens of thousands of years of reproductive isolation for distinct lineages of mitochondrial DNA to arise; significant morphological, ecological, and behavioral differences considered characteristic of separate species may arise over that length of time as well.
3. The paradoxical observation of large differences between species (indicating steady change) and small differences within (indicating change is constrained) implies that the pool of variants within a species changes steadily over evolutionary time scales. Like influenza virus, which regularly produces new variants that replace last year’s strains, the DNA sequences within breeding populations are continuously evolving, so that reproductive isolation over a sufficient period of time inevitably leads to genetic divergence. There may be morphologic stasis but there is no genetic stasis.
4. The usual absence of multiple lineages (with say >1% divergence in coding mtDNA) within breeding populations implies selection against hybrids and their offspring.
Now for some complex real data that challenge this simple model! In Proc R Soc B August 2007, researchers report on “Limited performance of DNA barcoding in a diverse community of tropical butterflies”. Elias and colleagues examined COI barcode region mtDNA sequences in 353 specimens from 57 species of ithomiine butterflies, most from 2 study sites in eastern Ecuador. Ithomiines are a tropical subfamily of approximately 360 species, virtually all of which are part of dizzyingly complex “rings” of Mullerian mimicry (all species distasteful) in which multiple species, some only distantly related, have nearly identical morpholgy. There is often marked geographic variation within what are considered single species such that different regional forms participate in different rings. For more appreciation, there are gorgeously illustrated research and other sites on ithomiines and other Mullerian mimics.
This exemplary study helps demonstrate the power of analyzing a standardized region, ie DNA barcoding, as their findings can be directly compared to results in other studies. In NJ analysis using the 273 study site specimens, the authors found that 44 of 57 (77%) of species formed well-supported (>50%) clusters. When sequences from non-local specimens were added to the analysis, and considering only species with more than one congener and with local and non-local sequences, 28 of 41 species (68%) formed distinct clusters. So one might mark down this group as challenging for DNA barcode approach to species identification.
One question is whether genetic diversity is more finely divided than current taxonomy recognizes. Differences within species sampled at distant geographic sites were as high as 8.5%, which the authors view as expected variation for tropical species with large census population sizes. Is this correct? Do larger populations support greater mitochondrial variation? According to report last year by Bazin et al Science 312:570 April 2006, the answer is no, but this conclusion seems not yet widely embraced. Following Bazin et al and the model outlined above, I suggest the genetically divergent forms reflect reproductively isolated allopatric populations and some might turn out to represent different species.
On the other end, some species had nearly identical COI sequences. Are these young species? The authors helpfully analyzed nuclear gene EF-1 alpha for most specimens and state that the nuclear gene sequence improved species-level identifications compared to mtCOI. On my inspection the published tree shows a similar overlap of EF-1 alpha gene sequences, which together with COI data suggests these are very closely-related young species. Recent work by some of the same authors Nature 441:868 14 June 2006 shows new species formation in just 3 generations in related Heliconidae butterflies through hybridization, so perhaps there are mechanisms that enable very rapid emergence of distinctive forms within these butterflies. There are presumably swarms of populations within many species that are distinctive in one form or another.
As this study shows, comparing relative and absolute differences in a standardized gene region is a useful approach for exploring the genetics of biodiversity. DNA barcode data sets can help address the question of whether population size influences mitochondrial sequence variation, and in turn the answer will help in understanding the patterning of genetic diversity among and within species. I look forward to more data on ithomiines and their relatives!
APPEA address published
The Australian Petroleum Production and Exploration Association has published Jesse’s April 2007 Plenary address “The Future Environment for the Energy Business†in the 2007 APPEA Journal. A key theme of the paper is responsibility of the oil and gas industries for protection of marine life.
Scanning mosquito barcodes to help solve disease mystery
 What limits Japanese encephalitis virus (JEV) to its current range? JEV is a mosquito-transmitted flavivirus related to yellow fever and West Nile viruses that causes approximately 40,000 human cases annually in SE Asia. Although regular epidemics occur in islands off Papua New Guinea as close as 70 km to Australia and the major JEV vector in Papua New Guinea (PNG), Culex annulirostris, is found throughout Australia, there have only been sporadic cases in Australia and the disease has not become established there.
What limits Japanese encephalitis virus (JEV) to its current range? JEV is a mosquito-transmitted flavivirus related to yellow fever and West Nile viruses that causes approximately 40,000 human cases annually in SE Asia. Although regular epidemics occur in islands off Papua New Guinea as close as 70 km to Australia and the major JEV vector in Papua New Guinea (PNG), Culex annulirostris, is found throughout Australia, there have only been sporadic cases in Australia and the disease has not become established there.
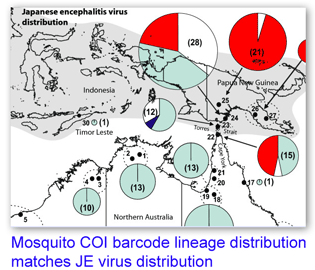 In 29 June 2007 BMC Evol Biol researchers analyzed mitochondrial COI and nuclear ITS 1 sequences in 273 mosquitos identified as Culex annulirostris or its close relatives Cx. palpalis and Cx. sitiens, collected at 30 locations in Australia and Papua New Guinea. Hemmerter et al found that 10% of morphological identifications were incorrect, based on ITS 1 sequences, and there was “100% agreement between the ITS 1 diagnostic and the COI sequence grouping of Culex spp.” Bayesian phylogenetic trees with COI showed “distinct geographically-structured lineages” (ie possible cryptic species) within the vector species Culex annulirostris, and two of the four Cx. annulirostris lineages are restricted to PNG, with a southern limit at the top of Australia’s Cape York peninsula, “which correlates exactly with the current southern limit of JEV activity”. Analysis of blood meals reveals the Australian Cx. annulirostris feed mainly on marsupials (PNG lineages feed on wild pigs which are the primary JEV reservoir), and laboratory studies indicate Australian Cx. annulirostris is an inefficient vector for JEV. As the authors note, it seems likely these genetically and biologically distinct lineages are likely different species.
In 29 June 2007 BMC Evol Biol researchers analyzed mitochondrial COI and nuclear ITS 1 sequences in 273 mosquitos identified as Culex annulirostris or its close relatives Cx. palpalis and Cx. sitiens, collected at 30 locations in Australia and Papua New Guinea. Hemmerter et al found that 10% of morphological identifications were incorrect, based on ITS 1 sequences, and there was “100% agreement between the ITS 1 diagnostic and the COI sequence grouping of Culex spp.” Bayesian phylogenetic trees with COI showed “distinct geographically-structured lineages” (ie possible cryptic species) within the vector species Culex annulirostris, and two of the four Cx. annulirostris lineages are restricted to PNG, with a southern limit at the top of Australia’s Cape York peninsula, “which correlates exactly with the current southern limit of JEV activity”. Analysis of blood meals reveals the Australian Cx. annulirostris feed mainly on marsupials (PNG lineages feed on wild pigs which are the primary JEV reservoir), and laboratory studies indicate Australian Cx. annulirostris is an inefficient vector for JEV. As the authors note, it seems likely these genetically and biologically distinct lineages are likely different species.
One limitation of this study is that the COI region analyzed does not match the COI barcode region. By my analysis the 538-bp fragment analyzed in this study starts at position 359 in COI. As the defined COI barcode region is 648 bp starting at position 58, there is only 289 bp overlap between the sequences in this study and COI barcodes. It appears generally straightforward to amplify COI barcodes from insects including mosquitos, so I hope the next study on genetic differences in human disease vectors will amplify the COI barcode region, as that will enable linking the results to the growing DNA barcode library, amplifying the power of the research itself.
I conclude that routine application of standardized genetic testing, ie DNA barcoding, will help in understanding the distribution of mosquito biodiversity, with implications for human health.
Marine barcode of life initiative joins web panoply
In July 2007 the Marine Barcode of Life initiative (MarBOL) surfaced at www.marinebarcoding.org. MarBOL is “an international initiative to enhance our capacity to identify marine life by utilizing DNA barcoding”. It is an offspring of the Census of Marine Life (CoML), a ten-year initiative to assess and explain the diversity, distribution, and abundance of marine life in the oceans and the DNA barcode initiative.
The target list for MarBOL includes the diverse invertebrates that inhabit the oceans, as well as marine mammals, fish, and birds. MarBOL will be compiling barcodes collected through CoML projects, including those focused on marine zooplankton (CMarZ), pelagic animals (TOPP), nearshore environments (NaGISA), reefs (CReefs), continental shelves (COMARGE), seamounts (CenSeam), deep water vents (ChEss), abyssal plains (CeDAMar), Arctic Ocean (ArcOD), Antarctic Ocean (CAML), northern Mid-Atlantic ridge (MAR-ECO), Gulf of Maine (GoMA), northeastern Pacific continental shelf (POST), and perhaps even marine microbes (IcoMM)! The project will also utilize barcodes collected by ongoing barcoding initiatives on fish, birds, and sponges.
CoML Bluefin Tuna
The abundance of bluefin tuna in the North Atlantic are the subject of a Census of Marine Life press release . The original tuna papers as well as tuna images and video are are on the CoML Portal.
EOL Videos
The 8-9 May 2007 public launch of the Encyclopedia of Life included short appearances by Jesse on Reuters television and on the CBC evening news.
Part stands for the whole
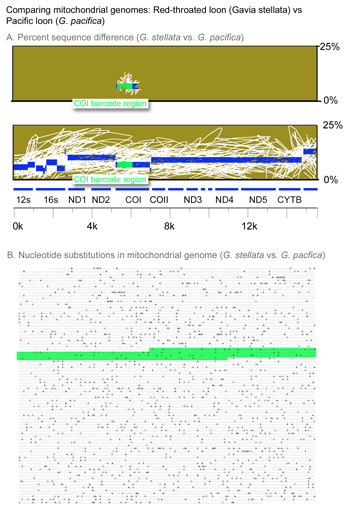
A synecdoche is a figure of speech in which a part stands for the whole, or the whole stands for a part. Taking the first, we might consider a DNA barcode as a synecdoche, in which the short barcode gene fragment stands for whole genome. As in the figure, a COI barcode usually encapsulates the differences found elsewhere in the mitochondrial genome. Because COI barcodes generally capture the discontinuities we recognize as species, we can surmise that differences in this short mitochondrial gene fragment usually reflect differences in the nuclear genome. More study of variation within and among species will help understand why differences in mitochondrial and nuclear genomes appear inextricably linked.
for larger version click here
