 What lives in soil? In August 2009 Pesq Agropec Bras (open access) an international cohort of 10 researchers from Canada, France, US, Taiwan, and Russia examine prospects for speeding assessment of soil animal diversity with COI DNA barcoding. As test sets, Rougerie and colleagues explore taxonomic and sequence diversity in earthworms and collembolans (springtails). These two groups comprise a similar number of named species (earthworms, 6000; springtails, 7900), but these totals likely underestimate true diversity, particularly for springtails, which are tiny (0.2 – 6.0 mm), challenging collecting and morphology.
What lives in soil? In August 2009 Pesq Agropec Bras (open access) an international cohort of 10 researchers from Canada, France, US, Taiwan, and Russia examine prospects for speeding assessment of soil animal diversity with COI DNA barcoding. As test sets, Rougerie and colleagues explore taxonomic and sequence diversity in earthworms and collembolans (springtails). These two groups comprise a similar number of named species (earthworms, 6000; springtails, 7900), but these totals likely underestimate true diversity, particularly for springtails, which are tiny (0.2 – 6.0 mm), challenging collecting and morphology.
For earthworms, the researchers analyzed COI sequences from 457 specimens collected in 13 countries around the world (including North America, South America, Caribbean, Europe, Middle East, Southeast Asia, and Australia); these represented at least 49 genera in 8 families. 87 species were identified by morphology, representing about 1/2 of specimens; the remainder, mostly those from Philippines and Brazil, could not be identified to species. Applying a threshold approach, the researchers found 192 (10% cutoff) and 211 (4% cutoff) genetic clusters, including two or more divergent clusters in 13 (15%) of named species. None of species showed sequence sharing or overlaps.
For springtails, 695 specimens from a similar global distribution of sites were analyzed, representing 88 genera in 16 familes; only 44 species were formally identified with morphologic characters, consistent with the “difficult and poorly known taxonomy of these organisms.” Of note, the authors report that “a specific protocol was developed for [collembolans] so that voucher specimens could be recollected after DNA extraction and thus be used for further morphological examination.” Sequence comparisons showed a “typical…bimodal distribution of intra- versus interspecific divergences such as the one also reported in earthworms.” Applying distance thresholds as above gave 215 (10% cutoff) and 227 (4% cutoff) genetic clusters. In conclusion, efficient assessment of soil animal diversity calls out for DNA barcoding.
On a separate note, soil animals may be useful for understanding mitochondrial evolution. Even factoring in species diversity, these animals appear to have enormous population sizes, given densities up to 4 x 103 earthworms/m2 and 1.8 x 106 collembolans/m3, yet show typical bimodal pattern of intra- << inter-specific variation noted above. Along these lines, in November 2009 Nature Nick Lane explores whys of mitochondrial evolution and speciation, including a possible “radically new picture of mitochondrial genes being tightly regulated by selection.” Stay tuned!
 What lives in soil? In
What lives in soil? In 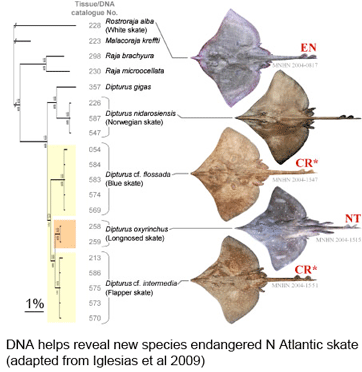 As the researchers report, this taxonomic oversight obscured the disappearance of one species, the flapper skate (D. cf. intermedia) because it was confused with the less threatened blue skate (D. cf. flossada). Iglesias and colleagues marketplace survey revealed additional sources of confusion. They analyzed 4,110 skates landed over a 2 year period from 103 fishing cruises in four main French ports by 41 different French commercial trawlers, and found that five skate species (included the two named above) from two genera are variously lumped together under just two marketplace names, the aforementioned “European common skate (D. batis)” and “longnose skate (D. oxyrinchus);” according to their analysis the latter species, formerly common, is also locally extirpated, and most specimens with this name represent other species.
As the researchers report, this taxonomic oversight obscured the disappearance of one species, the flapper skate (D. cf. intermedia) because it was confused with the less threatened blue skate (D. cf. flossada). Iglesias and colleagues marketplace survey revealed additional sources of confusion. They analyzed 4,110 skates landed over a 2 year period from 103 fishing cruises in four main French ports by 41 different French commercial trawlers, and found that five skate species (included the two named above) from two genera are variously lumped together under just two marketplace names, the aforementioned “European common skate (D. batis)” and “longnose skate (D. oxyrinchus);” according to their analysis the latter species, formerly common, is also locally extirpated, and most specimens with this name represent other species.