NYC/NJ Aquatic Environmental DNA (eDNA) Project
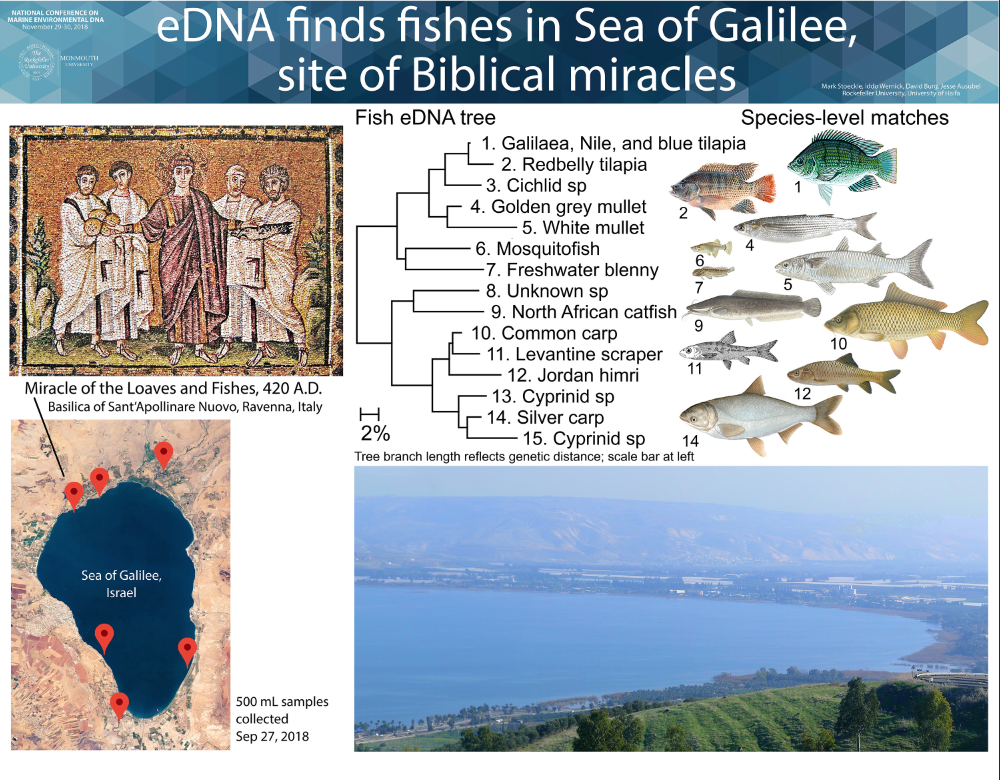
We aim to help advance eDNA for monitoring ocean life. Our research focuses on eDNA assessment of presence and abundance of marine fish in New York and New Jersey. We have also applied eDNA to freshwater fish, marine mammals, and terrestrial vertebrates, at diverse sites ranging from Sea of Galilee in Israel to twilight zone ocean at edge of eastern US continental shelf.
Why eDNA? Ocean life is largely hidden from view. Traditional surveys for marine animals are costly, time- and capital-intensive, needing special equipment and trained personnel. Surveys are relatively sparse, and many regions remain unexplored.
eDNA offers a low-cost, non-destructive complement to traditional surveys, with significant added value in otherwise difficult-to-survey environments, for protected species, and if pandemics or other events curtail routine surveys. eDNA will help monitor impacts of human activities, weather and climate, conservation efforts, and will aid research and exploration.
Optimizing eDNA methods for metabarcoding marine fish. eDNA metabarcoding uses PCR primers that amplify the DNA of multiple species in a taxonomic group, e.g., vertebrates. The amplified DNA is analyzed by high-throughput NextGen sequencing, and the resulting “reads” are sorted and matched to a reference library of sequences from identified fish specimens. We have shown that current laboratory protocols optimize sensitivity and reproducibility, especially for more abundant fish populations (ICES J Mar Sci 2022a). We conclude that eDNA rarity poses the main challenge to current methods.
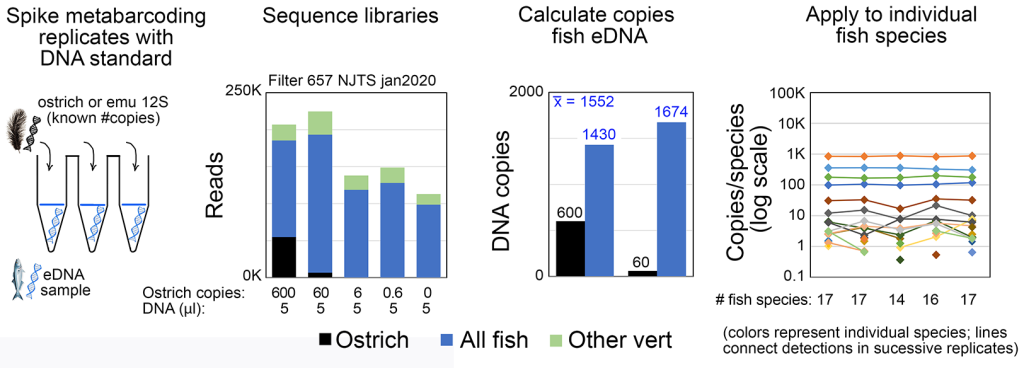
eDNA metabarcoding has generally been considered best suited for presence/absence assessment rather than as a measure of abundance. We recently demonstrated that with certain primer sets, spiking metabarcoding PCRs with an exogenous standard (e.g., ostrich DNA as illustrated below) enables accurate measurement of eDNA copy number levels (ICES J Mar Sci 2022b). This opens the door to comparing fish abundance among species, by habitat and season, and monitoring response to restoration efforts. Our work in this area is currently supported by NOAA Award Number NA23OAR0110593.
_______________________________________________________________________________________
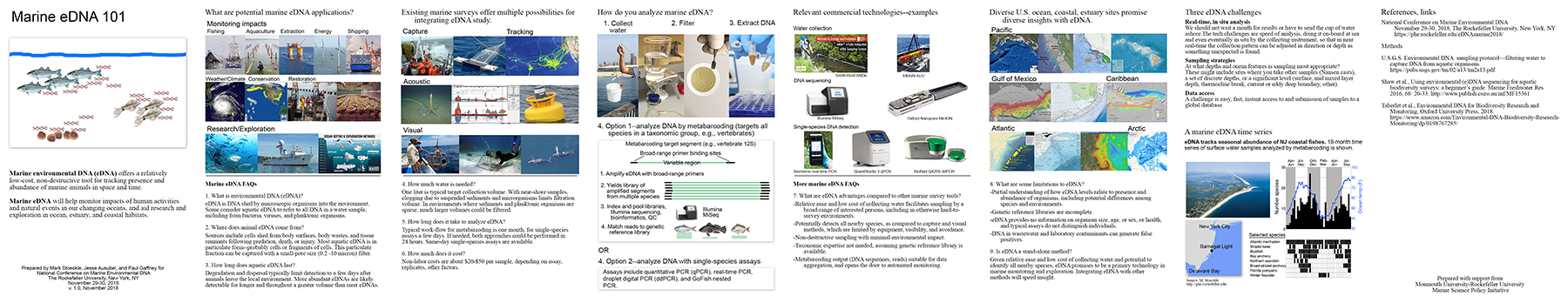
Marine eDNA 101 booklet pdf (618 KB)
_______________________________________________________________________________________

In collaboration with Monmouth University and New Jersey Department of Environmental Protection, we conducted one of the largest comparisons of eDNA and bottom trawling to date. We found the two technologies were largely concordant on marine fish diversity, seasonality, and relative abundance (link to article ICES J Marine Sci 2021, also see Science online coverage by Erik Stokstad).
______________________________________________________________________________________
Together with Monmouth University, we hosted the first National Conference on Marine Environmental DNA, November 29-30, 2018. The Conference included approximately 100 American ocean scientists and associated stakeholders, including representatives from academe, federal, state, and local governments, non-governmental organizations concerned with marine environment, and the private sector. The strong sense of the meeting was “eDNA works–let’s get going.” The Conference Final Report and press release summarize the conference proceedings and outline concrete steps forward.
The Conference attracted coverage in Science online and print, and National Geographic online, reflecting the excitement around marine eDNA for monitoring and exploring ocean life.
_______________________________________________________________________________________
More eDNA explorations
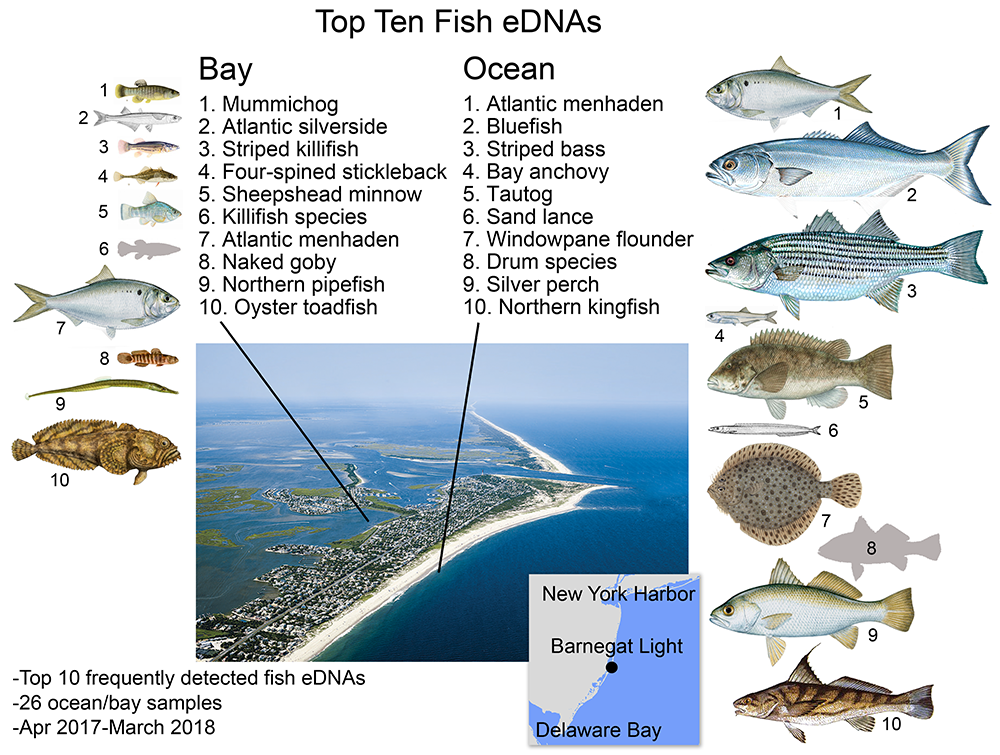
Bay fish eDNA in bay, ocean fish eDNA in ocean
Contact: Mark Stoeckle mark.stoeckle@rockefeller.edu Site updated November 2020
More aquatic eDNA projects
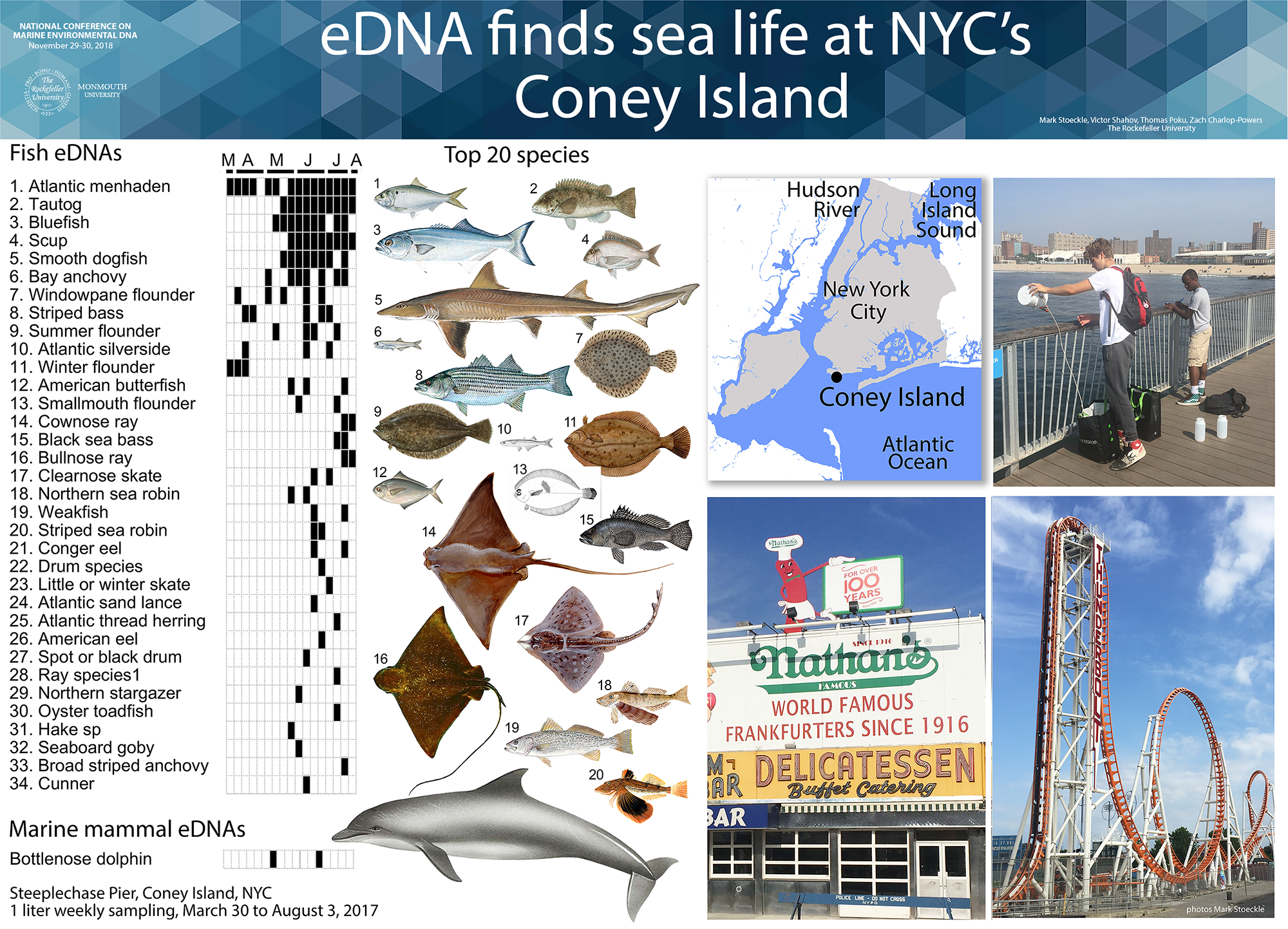
2015 NYC Marine eDNA survey 2015 eDNA NYC Central Park
Publications
Mark Y. Stoeckle, Jesse H. Ausubel, Michael Coogan. 12S gene metabarcoding with DNA standard quantifies marine bony fish environmental DNA, identifies threshold for reproducible detection, and overcomes distortion due to amplification of non-fish DNA (PDF). , Environmental DNA, (eds.). DOI: 10.1002/edn3.376 (11 November): 2022
Stoeckle MY, Adolf J, Ausubel JH, Charlop-Powers Z, Dunton K, Hinks G. Current laboratory protocols for detecting fish species with environmental DNA optimize sensitivity and reproducibility, especially for more abundant populations [open access external link]. ICES J Marine Sci, 2022.
Stoeckle MY, Adolf J, Charlop-Powers Z, Dunton K, Hinks G, VanMorter SM. Trawl and eDNA assessment of marine fish in coastal New Jersey, USA [open access external link]. ICES J Marine Sci, 2021.
Improved Environmental DNA Reference Library Detects Overlooked Marine Fishes in New Jersey, United States [open access external link]. Frontiers in Marine Science 7 (226): 2020
Stoeckle MY, Mishu M, Charlop-Powers Z. GoFish: a versatile strategy for nested PCR environmental DNA assays for marine vertebrates [open access external link]. PLOS ONE 2018:e0198717.
Stoeckle MY. Fishing for DNA: free-floating eDNA identifies presence and abundance of ocean life [open access external link]. The Conversation, April 12, 2017.
Stoeckle MY, Soboleva L, Charlop-Powers Z. Aquatic environmental DNA detects seasonal fish abundance and habitat preference in an urban estuary [open access external link]. PLOS ONE 2017: e0175186.
About the Bar Code of Life site
This web site is an outgrowth of
the Taxonomy, DNA, and Barcode of Life meeting held at Banbury
Center, Cold Spring Harbor Laboratory, September 9-12, 2003.
It is managed by Mark Stoeckle at the Program
for the Human Environment (PHE) at The Rockefeller University.
Contact: mark.stoeckle@rockefeller.edu
About the Program
for the Human Environment
The involvement of the Program for the Human Environment in DNA
barcoding dates to Jesse Ausubel's attendance in February 2002
at a conference in Nova Scotia organized by the Canadian Center
for Marine Biodiversity. At the conference, Paul Hebert
presented for the first time his concept of large-scale DNA
barcoding for species identification. Impressed by the
potential for this technology to address difficult challenges
in the Census of Marine Life, Jesse agreed with Paul on
encouraging a conference to explore the contribution
taxonomy and DNA could make to the Census as well as other large-scale
terrestrial efforts. In his capacity as a Program Director of
the Sloan Foundation, Jesse turned to the Banbury Conference
Center of Cold Spring Harbor Laboratory, whose leader Jan
Witkowski prepared a strong proposal to explore both the
scientific reliability of barcoding and the processes that
might bring it to broad application. Concurrently, PHE
researcher Mark Stoeckle began to work with the Hebert lab on
analytic studies of barcoding in birds. Our involvement in
barcoding now takes 3 forms: assisting the organizational
development of the Consortium for the Barcode of Life and the
Barcode of Life Initiative; contributing to the scientific
development of the field, especially by studies in birds, and
contributing to public understanding of the science and
technology of barcoding and its applications through improved
visualization techniques and preparation of brochures and other
broadly accessible means, including this website. While the
Sloan Foundation continues to support CBOL through a grant to
the Smithsonian Institution, it does not provide financial
support for barcoding research itself or support to the PHE for
its research in this field.