Simplified DNA barcode recipe-skip step one
Thursday, April 15th, 2010
It used to be standard practice to shave the area around the incision before surgery, as it was thought that hair harbored bacteria that would cause wound infection. Beginning in the 1970s, doctors found this was unnecessary, and in fact was associated with higher incidence of post-operative infection. This history comes to mind in reading March 2010 BioTechniques report by researchers from University of Guelph demonstrating that DNA suitable for PCR and sequencing can be obtained simply by leaving specimens in alcohol overnight!
 Of the three steps required to get from a specimen to a DNA barcode, namely DNA isolation, PCR (polymerase chain reaction), and sequencing, the first step is the most labor intensive and hardest to automate. Numerous protocols/kits have been developed to optimize DNA isolation from various types of specimens, such as plant vs animal tissues. As described by the Guelph researchers, “these procedures force cells to release their DNA via physical pertubation and/or chemical treatment, which is then followed by a clean-up procedure in which unwanted cellular compoents are separated from the DNA.” The researchers “hypothesized that a small amount of DNA leaks from the tissue into the preservation solution (usually ethanol), and that this DNA was amplifiable using a standard PCR protocol.” To start, they analyzed Monte Alban mescal, which is sold with a “worm” (a caterpillar of the agave moth, Hypopta agavis) in each bottle. They evaporated 50 mL mescal, re-dissolved the residue in water, applied this to a Qiagen MinElute spin column, resuspended the product in 50 μL water, and used 2 μL of resulting solution in a standard 25 μL PCR reaction, with successful amplification and sequencing of 130 base mini-barcode of COI. This case was presumably challenging as mescal is only 40% ethanol and contains a variety of material that might inhibit PCR. In subsequent tests, 1 mL of 95% ethanol used to preserve specimens was evaporated, resuspended in 30 μL of water without column purification, and 2 μL used for PCR.
Of the three steps required to get from a specimen to a DNA barcode, namely DNA isolation, PCR (polymerase chain reaction), and sequencing, the first step is the most labor intensive and hardest to automate. Numerous protocols/kits have been developed to optimize DNA isolation from various types of specimens, such as plant vs animal tissues. As described by the Guelph researchers, “these procedures force cells to release their DNA via physical pertubation and/or chemical treatment, which is then followed by a clean-up procedure in which unwanted cellular compoents are separated from the DNA.” The researchers “hypothesized that a small amount of DNA leaks from the tissue into the preservation solution (usually ethanol), and that this DNA was amplifiable using a standard PCR protocol.” To start, they analyzed Monte Alban mescal, which is sold with a “worm” (a caterpillar of the agave moth, Hypopta agavis) in each bottle. They evaporated 50 mL mescal, re-dissolved the residue in water, applied this to a Qiagen MinElute spin column, resuspended the product in 50 μL water, and used 2 μL of resulting solution in a standard 25 μL PCR reaction, with successful amplification and sequencing of 130 base mini-barcode of COI. This case was presumably challenging as mescal is only 40% ethanol and contains a variety of material that might inhibit PCR. In subsequent tests, 1 mL of 95% ethanol used to preserve specimens was evaporated, resuspended in 30 μL of water without column purification, and 2 μL used for PCR.
By evaporating 1 mL of ethanol in which specimens had been stored overnight (out of 2 mL total ethanol volume) and re-suspending residue in water, Shokralla and colleagues amplified and sequenced 130-base and 650-base fragments of COI and 1100-base fragment of 28s RNA from 25 whole insect specimens (mayflies, caddisflies; 1 gave COI only) and rbcL from 45 plant specimens (0.5 mm leaf samples). They also obtained COI sequences by sampling 1 mL of ethanol solution from 7 insect specimens stored at room temperature for 7 to 10 years. The researchers note this approach could facilitate for “high-throughput” analyses, as it involves liquid handling which is easy to automate, avoids destructive sampling, and could be used even when “there is simply no sample left for further analysis.” They conclude with a caution about “field sampling procedures that include placing mixtures of specimens in an ethanol jar” as this “may increase the chance of cross-contamination.”
The remarkably simple procedure reported by Shokralla and colleagues offers benefits to many persons who want to get DNA barcode identifications. I look forward to applications of this method in research and commercial laboratories, classrooms, and perhaps kitchens!
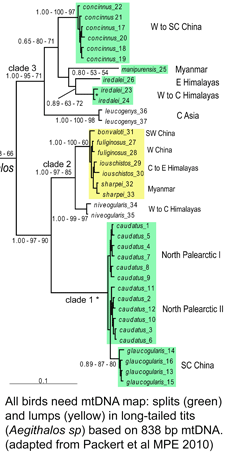 Although birds have been studied in more detail than any other large group of animals, mtDNA continues to reveal many overlooked species, such that named taxa turn out to be comprised of two or more distinct species. These revisions include some very familiar birds, e.g., Canada Goose, which was recently recognized as comprising two species, Cackling Goose (B. hutchinsii) and Canada Goose (B. canadensis) (
Although birds have been studied in more detail than any other large group of animals, mtDNA continues to reveal many overlooked species, such that named taxa turn out to be comprised of two or more distinct species. These revisions include some very familiar birds, e.g., Canada Goose, which was recently recognized as comprising two species, Cackling Goose (B. hutchinsii) and Canada Goose (B. canadensis) (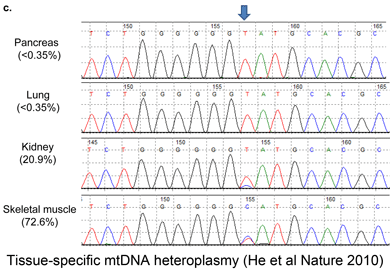 To establish a cutoff for artefactual errors due to PCR and/or sequencing, a control comparison with amplified nuclear DNA was performed, which yielded an average of 0.058% (SD 0.057%) mutations per base and a maximum of 0.82% mutations. He and colleagues used a “very conservative assumption that all variants in excess of twice this value (1.6%) represented true heteroplasmies rather than sequencing artefacts.” Now to some results! The researchers detected “28 homoplasmic alleles and 8 heteroplasmic alleles in this sample of normal colonic mucosa.” Here “homoplastic” refers to differences from the reference human mtDNA sequence (
To establish a cutoff for artefactual errors due to PCR and/or sequencing, a control comparison with amplified nuclear DNA was performed, which yielded an average of 0.058% (SD 0.057%) mutations per base and a maximum of 0.82% mutations. He and colleagues used a “very conservative assumption that all variants in excess of twice this value (1.6%) represented true heteroplasmies rather than sequencing artefacts.” Now to some results! The researchers detected “28 homoplasmic alleles and 8 heteroplasmic alleles in this sample of normal colonic mucosa.” Here “homoplastic” refers to differences from the reference human mtDNA sequence ( In
In  DNA helps answer the origin of infectious diseases: are cases sporadic events or part of larger epidemic, such as the
DNA helps answer the origin of infectious diseases: are cases sporadic events or part of larger epidemic, such as the 
 In terms of citizen participation, the MPG story suggests expanding opportunities for biological research that harnesses the skill and energy of non-professionals, a step beyond the successful
In terms of citizen participation, the MPG story suggests expanding opportunities for biological research that harnesses the skill and energy of non-professionals, a step beyond the successful  Herbal products make a compelling case for DNA-based identification–how else to recognize dried bits of roots, leaves, stems, bark, and flowers from a multitude of species? In
Herbal products make a compelling case for DNA-based identification–how else to recognize dried bits of roots, leaves, stems, bark, and flowers from a multitude of species? In  This study demonstrates advantages of DNA barcoding approach for plant identification. Of course, there is already a lot of interest in DNA identification of herbal plants in general and Dendrobium orchids in particular. For example, I found over a dozen articles describing DNA methods for distinguishing Dendrobium sp. However, the methods described are limited to identifying species in this one genus, which means one has to have a pretty good idea what the specimen is before applying DNA testing! This highlights the essential advantage of barcoding–a standardized approach can be applied to any unknown, and makes feasible creation of a comprehensive reference library.
This study demonstrates advantages of DNA barcoding approach for plant identification. Of course, there is already a lot of interest in DNA identification of herbal plants in general and Dendrobium orchids in particular. For example, I found over a dozen articles describing DNA methods for distinguishing Dendrobium sp. However, the methods described are limited to identifying species in this one genus, which means one has to have a pretty good idea what the specimen is before applying DNA testing! This highlights the essential advantage of barcoding–a standardized approach can be applied to any unknown, and makes feasible creation of a comprehensive reference library. In forensic investigation, insect evidence helps date the time of death, as the various species that colonize corpses exhibit different stages of development according to time and temperature. Determining the post-mortem interval (PMI) rests on accurate species identification, including of immature forms. In
In forensic investigation, insect evidence helps date the time of death, as the various species that colonize corpses exhibit different stages of development according to time and temperature. Determining the post-mortem interval (PMI) rests on accurate species identification, including of immature forms. In 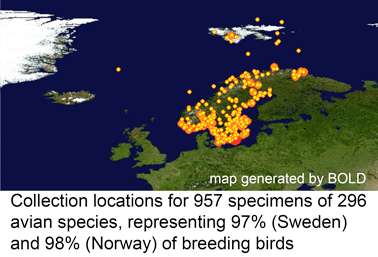 In
In 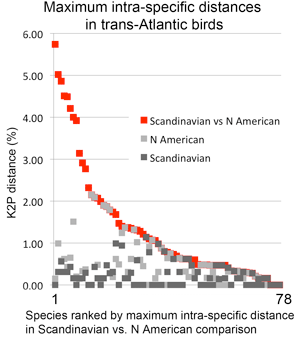 In my view, this paper demonstrates that a survey approach produces a high level of discovery and hypothesis-generating, and leads me to question how well we understand diversity in birds, which are generally considered the taxonomically best-known large group of animals. Many of the species in the present study have been known to science for over 250 years, are resident in densely-settled, scientifically-advanced regions, and yet Johnsen and colleagues demonstrate hidden diversity. In 1946, Ernst Mayr compiled a
In my view, this paper demonstrates that a survey approach produces a high level of discovery and hypothesis-generating, and leads me to question how well we understand diversity in birds, which are generally considered the taxonomically best-known large group of animals. Many of the species in the present study have been known to science for over 250 years, are resident in densely-settled, scientifically-advanced regions, and yet Johnsen and colleagues demonstrate hidden diversity. In 1946, Ernst Mayr compiled a  temporary placeholders is “taxon label”). As they note, there are many provisional names in GenBank (e.g. Ocyptamus sp. MZH S143_2004), so this is not a change in usual practice, except that the format of provisional names is standardized. As a starting point, Schindel and Miller propose a scheme developed by Council of the Heads of Australian Herbariums (CHAH) and recommend review by
temporary placeholders is “taxon label”). As they note, there are many provisional names in GenBank (e.g. Ocyptamus sp. MZH S143_2004), so this is not a change in usual practice, except that the format of provisional names is standardized. As a starting point, Schindel and Miller propose a scheme developed by Council of the Heads of Australian Herbariums (CHAH) and recommend review by