Avian catalogue still incomplete
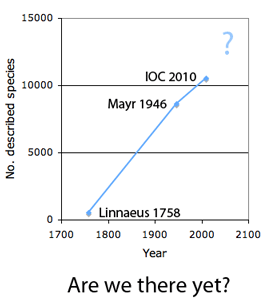 How many birds in the world? In the tenth edition of Systema Naturae (1758) (copy in US Library of Congress can be viewed or downloaded here, thanks to Biodiversity Heritage Library), Linnaeus listed 564 species collected from all over the world. In 1935, Ernst Mayr estimated 8,500 world birds, and counted more precisely in 1946, arriving at a total of 8,616 species (Auk 63:64-67). Mayr judged “this figure is probably within five per cent, and certainly within ten per cent, of the final figure” and predicted “whatever changes may occur in the future will be due primarily to taxonomic revaluations, that is to shifts from specific to subspecies status and vice versa.” As of today, the IOC World Bird List v2.4 names 10,386 species, plus another 139 accepted or proposed splits, altogether about 20% above Mayr’s 50 year-old estimate.
How many birds in the world? In the tenth edition of Systema Naturae (1758) (copy in US Library of Congress can be viewed or downloaded here, thanks to Biodiversity Heritage Library), Linnaeus listed 564 species collected from all over the world. In 1935, Ernst Mayr estimated 8,500 world birds, and counted more precisely in 1946, arriving at a total of 8,616 species (Auk 63:64-67). Mayr judged “this figure is probably within five per cent, and certainly within ten per cent, of the final figure” and predicted “whatever changes may occur in the future will be due primarily to taxonomic revaluations, that is to shifts from specific to subspecies status and vice versa.” As of today, the IOC World Bird List v2.4 names 10,386 species, plus another 139 accepted or proposed splits, altogether about 20% above Mayr’s 50 year-old estimate.
As Mayr predicted, nearly all new birds represent “splits” of existing entities, often elevating described subspecies to species status. Mayr estimated about 28,500 “valid subspecies”–might these represent species? Most splits reflect, at least in part, newly discovered genetic differences in mtDNA. In 2004, Robert Zink examined in detail 41 widely-distributed N American birds, and found an average of 1.9 “historically significant units” per species, i.e., distinct mtDNA clusters, most or all of which likely represent distinct species (Proc R Soc Lond B 271:561). At the same time, he found over 90% of subspecies “lack the population genetic structure indicative of a distinct evolutionary unit.” I conclude that species-level avian taxonomy will benefit from a concerted effort to analyze mtDNA in all world birds, namely, All Birds Barcoding Initiative (ABBI). Large scale DNA barcoding surveys so far have found distinct mtDNA clusters in 4-24% of species (e.g., Kerr et al 2007, Kerr et al 2009, Johnsen et al 2010).
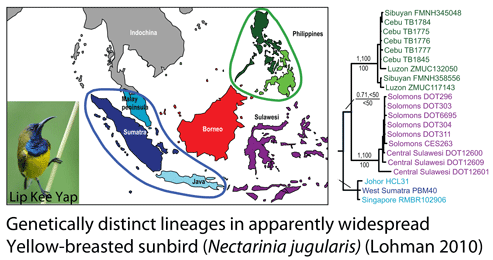 In some regions and categories of birds, the proportion of unrecognized species may be even higher. In August 2010 Biological Conservation researchers from 8 institutions in Southeast Asia and North America report on “cryptic genetic diversity” in non-migratory Philippine birds that are also apparently widespread in other Southeast Asian countries. Lohman and colleagues analyzed seven of the 72 non-migratory, non-endemic Philippine species in detail, represented by 210 tissue specimens (9-51 specimens/species), collected from 16 countries over 18 years by 54 collectors and held in 13 institutions!
In some regions and categories of birds, the proportion of unrecognized species may be even higher. In August 2010 Biological Conservation researchers from 8 institutions in Southeast Asia and North America report on “cryptic genetic diversity” in non-migratory Philippine birds that are also apparently widespread in other Southeast Asian countries. Lohman and colleagues analyzed seven of the 72 non-migratory, non-endemic Philippine species in detail, represented by 210 tissue specimens (9-51 specimens/species), collected from 16 countries over 18 years by 54 collectors and held in 13 institutions!
mtDNA analysis revealed genetically distinct clusters in all seven species (minimum Philippine/non-Philippine genetic distance 0.9-8.8% in COI, 2.1-9.4% in cytb). The researchers observe that using a “combination of monophyly, morphological distinctiveness as recognized by current subspecific taxonomy, and a 3% COI distance as a threshold for highlighting possible unrecognized species, six putatively new endemic Philippine species are revealed.” In addition to distinctness of Philippine forms, six of the seven species showed multiple (3-7) geographically-restricted lineages in Southeast Asia, at least some of which are likely to represent new species as well.
As Lohman and colleagues demonstrate, many of the tissue specimens needed to complete the census of world birds are already in museums, awaiting analysis. The world’s avian tissue collections comprise over 300,000 specimens representing over 7,000 species (Stoeckle and Winker Auk 2009), most of which, I surmise, have not been analyzed for any gene. DNA barcoding of existing avian tissue collections will likely lead to many discoveries.
This entry was posted on Wednesday, June 16th, 2010 at 12:20 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.
June 16th, 2010 at 1:09 pm
Thanks for the reference to the Biodiversity Heritage Library. One small correction, the copy of Systema Naturae (1758) referenced above is the copy from the Missouri Botanical Garden.