DNA plus database of sounds help reveal new bird species
 Birds are relatively large, conspicuous, vocal, and mostly diurnal creatures, making it relatively easy for humans to tell apart. Even so, new species continue to be discovered; these usually represent distinct forms within what were thought to be single species. In 27 February 2007 Mol Phylogenetics Evol, Arpad Nyari, University of Kansas, reports on molecular and vocal differentiation in a widespread South American songbird, the Thrush-like Schiffornis Schiffornis turdinus. S. turdinus is a “dull-colored, secretive bird distributed throughout Neotropical humid lowland forests from southeastern Mexico south to northern Bolivia and the Atlantic Forest of southeastern Brazil.” The thirteen recognized subspecies show “subtle differences in plumage hue and intensity, and body size”. How to sort out which forms might represent distinct species?
Birds are relatively large, conspicuous, vocal, and mostly diurnal creatures, making it relatively easy for humans to tell apart. Even so, new species continue to be discovered; these usually represent distinct forms within what were thought to be single species. In 27 February 2007 Mol Phylogenetics Evol, Arpad Nyari, University of Kansas, reports on molecular and vocal differentiation in a widespread South American songbird, the Thrush-like Schiffornis Schiffornis turdinus. S. turdinus is a “dull-colored, secretive bird distributed throughout Neotropical humid lowland forests from southeastern Mexico south to northern Bolivia and the Atlantic Forest of southeastern Brazil.” The thirteen recognized subspecies show “subtle differences in plumage hue and intensity, and body size”. How to sort out which forms might represent distinct species?
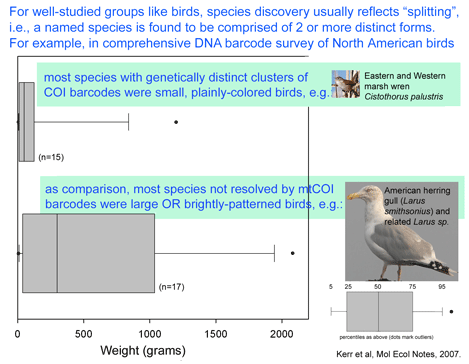
Nyari analyzed 38 individuals representing 10 of the 13 subspecies, plus 3 congeneric individuals of S. virescens or S. major. 8 of the 41 specimens were from University of Kansas, and the remainder were loaned from 6 museum collections in the US and Brazil, which highlights the distributed nature of avian tissue collections and the benefit of sharing resources. 2475 bp of mtDNA including COI barcode region, ND2, and cytochrome b were sequenced as molecular markers. Vocalizations were downloaded from Macaulay Library of Natural Sounds, Cornell University and spectrographs analyzed with RAVEN bioacoustic software which is provided on the Cornell site. The ability to combine these disparate data sets hints at power of making biological data widely available. Molecular analysis revealed 7 distinct geographically restricted clades, 5 of which had characteristic vocalizations. On this basis, S. turdina is recommended to be split into 5 species. When analyzed separately, the COI barcode region (615 bp) recovered the same 7 clades as the full 2475 bp. As in other studies, the branching order among clades and congenerics was better shown with the larger data set.
To my reading, this and other studies suggest that large-scale COI barcode screening of avian tissue collections will be a scientifically productive and efficient approach that will speed discovery of new bird species and advance understanding of avian diversity. As in Kerr et al’s recent study of North American birds, it seems likely that many or most of the new bird species awaiting discovery will be found among birds similarly inconspicuous as S. turdinus.
This entry was posted on Sunday, December 2nd, 2007 at 11:42 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.