Some Fret Over Exceptions to Barcoding
The springboard for a recent news@nature.com item by Hannah Hickey “Butterflies poke holes in DNA barcodes” is a report by Gompert et al in press in Mol. Ecology on genetic differences between two subspecies of Melissa blue butterfly, Lycaeides melissa melissa and L. m. samuelis. The latter subspecies is commonly known as “Karner blue” and is listed under the USA Endangered Species Act. Analysis of mitochondrial DNA revealed some populations of Karner blue have distinct COI sequences but those populations adjacent to the range of L. m. melissa subspecies do not. This result is not surprising. For one, DNA barcoding does not aim to separate subspecies. Subspecies are geographic variants within species whose differences shade into one another so it would be surprising if any single gene showed a sharp demarcation between populations. Most subspecies do not show diagnostic genetic differences, and when such differences are found, it has often led to proposals to elevate them to species status.
Regarding the utility of DNA barcoding, the findings with Melissa blues are unremarkable, as there are cases in all animal groups studied so far in which barcoding narrows identification to a few closely-related species, but no further. For example, see my earlier entry on comparing barcode performance. It may be helpful to point out that DNA barcoding is an instrument, not a theory. Cases of partial resolution do not “disprove” barcoding or invalidate its use. In fact, one application of DNA barcoding will be to quickly highlight such cases which may be biologically interesting as they likely represent recent speciation, ongoing hybridization, or synonymy.
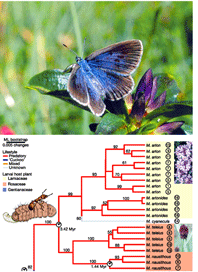
A more relevant Nature article that Ms. Hickey might have cited is Als et al study of Maculinea large blues, a related group morphologically similar, taxonomically confusing, and highly endangered butterflies. Large blues have “extraordinary parasitic lifestyles…later instars live in ant nests where they either devour the brood (predators), or are fed mouth-to-mouth by adult ants (cuckoos)”. Genetic analysis using mitochondrial and nuclear DNA uncovered numerous cryptic species with unsuspected host specificity, thereby both multiplying the challenge and providing the key to conservation, the need to conserve both ant hosts and butterflies.
As highlighted by the large blue study, the larger and more exciting challenge for biodiversity science will be how to incorporate the enormous number of genetically and biologically distinct forms whose discovery is facilitated by large-scale barcoding.
Photo of Rebel’s large blue Maculinea rebeli and phylogeny showing cryptic species among predatory Maculinea from Nature article by Als et al.
This entry was posted on Tuesday, March 28th, 2006 at 11:05 am and is filed under barcode performance, General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.