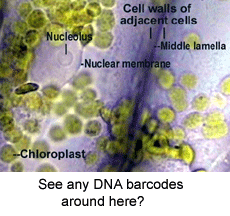
 Land plants challenge standardized DNA-based identification. Different groups of land plants are deeply divergent at the DNA level, yet there are relatively few sequence differences among closely-related species. Deep divergences make it difficult to design broad-range primers that amplify DNA from the many kinds of plants, and small differences among closely-related species mean longer sequences are needed to distinguish them. Plant mitochondrial genes including COI evolve too slowly to be useful. The best strategy appears likely to be a combination of 2 or 3 gene regions from the chloroplast genome. Chloroplasts are organelles which house the plants’ photosynthetic machinery and have their own genome, like mitochondria.
Land plants challenge standardized DNA-based identification. Different groups of land plants are deeply divergent at the DNA level, yet there are relatively few sequence differences among closely-related species. Deep divergences make it difficult to design broad-range primers that amplify DNA from the many kinds of plants, and small differences among closely-related species mean longer sequences are needed to distinguish them. Plant mitochondrial genes including COI evolve too slowly to be useful. The best strategy appears likely to be a combination of 2 or 3 gene regions from the chloroplast genome. Chloroplasts are organelles which house the plants’ photosynthetic machinery and have their own genome, like mitochondria.
In May 2007 Taxon, 19 researchers from 12 institutions in 7 countries (Brazil, Colombia, Denmark, Mexico, South Africa, U.K. and U.S.A.) report on tests of candidate barcode regions. Chase and co-investigators outline the rationale and results for selecting and testing potential land plant barcode regions. The finalists were winnowed down from more than 100 coding and non-coding regions in chloroplast DNA by testing 96 pairs of closely-related plant species to see which regions could be amplified and provide discrimination. Although the actual data are not shown in this short update, they summarize their results by proposing three chloroplast gene regions as a standard barcode for land plants: two coding regions, matK and rpoC1, and, either a third coding region, rpoB, or the non-coding psbA-trnH spacer region.
In June 2007 PLoS ONE, Kress and Erikcson, Smithsonian Institution, examine nine potential loci (8 plastid regions which includes the four final candidates in the Taxon paper, and nuclear gene ITS). In this analysis, as in Chase et al report, there are two steps: first, does the region amplify with a standard set of primers, and second, if so, does the sequence enable discrimination of closely-related species. In the 48 pairs of species examined, only two loci, trnH-psbA and rbcL-a exhibited more than 90% success with standard primers. Based on this admittedly small sample, the authors propose a “two-locus global DNA barcode for land plants” in which “rbcL-a provides a strong recognition anchor that will place an unidentified specimen into a family, genus, and sometimes species; the highly variable trnH-psbA spacer will futher narrow the corrrect species identification where rbcL-a lacks discrimination power.”
These are promising starts towards a standardized DNA barcode for land plants. More tests are needed, including analysis of variation within species, as both studies used single specimens for each target species.



 A bewildering array of morphologic nuances are needed to identify species. DNA sequences, aka DNA barcodes, can also be identifiers, with the advantage that anyone with the right device can name a specimen without having to consult an expert or search through taxonomic keys which are generally indecipherable to the non-specialist. As DNA barcode identifications become more common, will this mean a loss of biologically important knowledge? To my reading, the answer is no, as the subtle differences in specialized structures used to identify organisms generally provide little insight into how organisms live and what they do. Thus it is exciting that standardized COI DNA barcodes are being incorporated into species descriptions as this will help democratize access to species names and the biological knowledge they represent.
A bewildering array of morphologic nuances are needed to identify species. DNA sequences, aka DNA barcodes, can also be identifiers, with the advantage that anyone with the right device can name a specimen without having to consult an expert or search through taxonomic keys which are generally indecipherable to the non-specialist. As DNA barcode identifications become more common, will this mean a loss of biologically important knowledge? To my reading, the answer is no, as the subtle differences in specialized structures used to identify organisms generally provide little insight into how organisms live and what they do. Thus it is exciting that standardized COI DNA barcodes are being incorporated into species descriptions as this will help democratize access to species names and the biological knowledge they represent.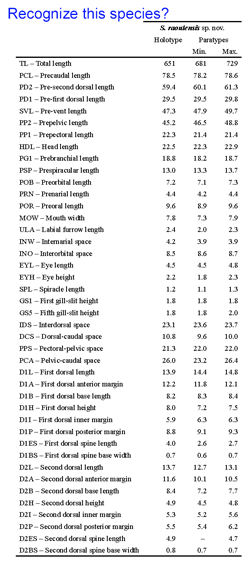 A
A 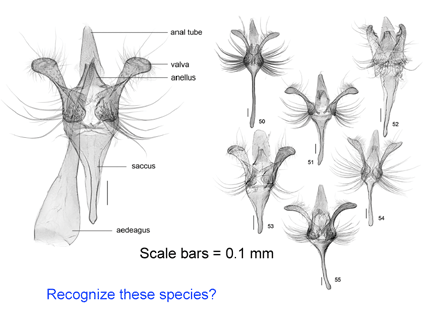
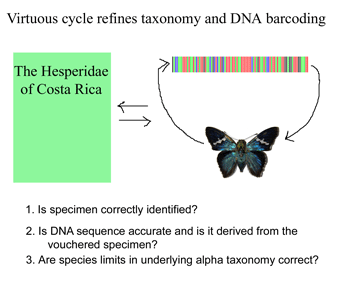 Like a map that is regularly updated, the reliability of DNA barcode databases will improve over time. To enable improvement, researchers have agreed to standardize on a particular region, to analyze multiple individuals from each species, and to revise DNA sequences and taxonomic labels as new information becomes available. By using specimens archived in museums, taxonomic identifications and DNA sequences can be re-checked. In
Like a map that is regularly updated, the reliability of DNA barcode databases will improve over time. To enable improvement, researchers have agreed to standardize on a particular region, to analyze multiple individuals from each species, and to revise DNA sequences and taxonomic labels as new information becomes available. By using specimens archived in museums, taxonomic identifications and DNA sequences can be re-checked. In  Growing data sets demonstrate DNA barcoding usually works, but why? Why does a very short stretch of DNA, such as a DNA barcode which usually represents less than one one-millionth of the genome, enable identification of most animal species? In computer language, Rod Page describes a DNA barcode as “
Growing data sets demonstrate DNA barcoding usually works, but why? Why does a very short stretch of DNA, such as a DNA barcode which usually represents less than one one-millionth of the genome, enable identification of most animal species? In computer language, Rod Page describes a DNA barcode as “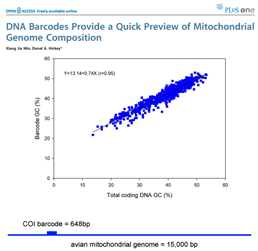 the DNA barcodes can provide a quick preview of the whole genome.” It will be of great interest to extend this analysis to compare mitochondrial barcodes to nuclear genomes; the general success of barcoding approach suggests there will be similarly close correlation.
the DNA barcodes can provide a quick preview of the whole genome.” It will be of great interest to extend this analysis to compare mitochondrial barcodes to nuclear genomes; the general success of barcoding approach suggests there will be similarly close correlation.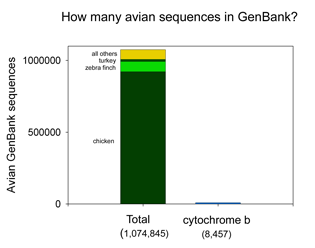 There are more than 1 million sequences in GenBank, but over 900,000 are from the Jungle Fowl (ie chicken, Gallus gallus), and another 85,000 from Zebra finch (Taeniopyga guttata) and Wild turkey (Meleagris gallopavo). That leaves about 67,000 sequences in total representing the rest of the approximately 10,000 species of world birds. According to Clements’ Birds of the World (including updates through 2006), there are 9,919 recognized species. The other world lists are very similar, and differ primarily in whether certain forms are recognized as species or subspecies and in assignment of generic names. I find it surprising there is not a single global taxonomic authority for bird species status, names, spelling, generic and family classification. As a comparison, medicine would be in great difficulty if there were not a single standard nomenclature for pathogenic bacteria.
There are more than 1 million sequences in GenBank, but over 900,000 are from the Jungle Fowl (ie chicken, Gallus gallus), and another 85,000 from Zebra finch (Taeniopyga guttata) and Wild turkey (Meleagris gallopavo). That leaves about 67,000 sequences in total representing the rest of the approximately 10,000 species of world birds. According to Clements’ Birds of the World (including updates through 2006), there are 9,919 recognized species. The other world lists are very similar, and differ primarily in whether certain forms are recognized as species or subspecies and in assignment of generic names. I find it surprising there is not a single global taxonomic authority for bird species status, names, spelling, generic and family classification. As a comparison, medicine would be in great difficulty if there were not a single standard nomenclature for pathogenic bacteria. 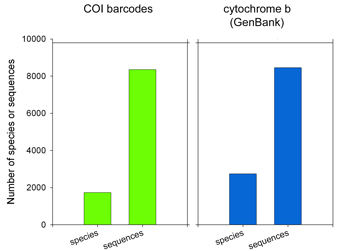 62,571 of the remaining 66,969 sequences are in the “CoreNucleotide” database (the others are unnamed genetic loci, either Expressed Sequence Tag (EST) or Genome Survey Sequence (GSS) records, and these will not be considered further here). Only 4,951 bird species are represented by any sequence (50% of world birds), and there are cytochrome b sequences for only 2,751 species (28% of world birds). Of species with cyt b sequences, 60% are represented by single sequences.
62,571 of the remaining 66,969 sequences are in the “CoreNucleotide” database (the others are unnamed genetic loci, either Expressed Sequence Tag (EST) or Genome Survey Sequence (GSS) records, and these will not be considered further here). Only 4,951 bird species are represented by any sequence (50% of world birds), and there are cytochrome b sequences for only 2,751 species (28% of world birds). Of species with cyt b sequences, 60% are represented by single sequences.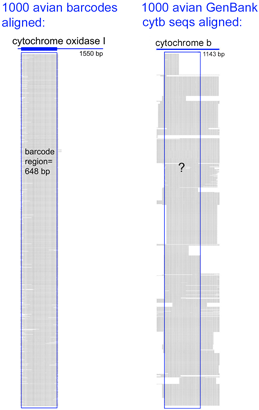 Virtues of the DNA barcode data set include that sequences are linked to vouchered museum specimens and their associated collecting data, sequence records include trace files to confirm sequencing accuracy, and most important all sequences can be directly compared because they derive from a standardized region. GenBank cyt b files include sequences of varying length and position along the gene. An alignment of 1000 avian COI barcodes and 1000 avian cyt b sequences hints at the power of a standardized approach.
Virtues of the DNA barcode data set include that sequences are linked to vouchered museum specimens and their associated collecting data, sequence records include trace files to confirm sequencing accuracy, and most important all sequences can be directly compared because they derive from a standardized region. GenBank cyt b files include sequences of varying length and position along the gene. An alignment of 1000 avian COI barcodes and 1000 avian cyt b sequences hints at the power of a standardized approach. A dream of many came to life this week with launch of
A dream of many came to life this week with launch of  The Scanning and Digitization Group will accelerate the work of the
The Scanning and Digitization Group will accelerate the work of the 
 In
In 
 The results show a “variegated picture of the taxonomic status of publicly indexed fungal sequences“. Taxonomic coverage is sparse: of the estimated 1.5 million fungi, less than 1% (9,684 species) are represented. Taxonomic data is lacking for many sequences (27% are not identified to species level), and most of the species-level identifications are unverifiable (82% are not linked to voucher specimens, 63% are not tagged with specimen country of origin, and 42% are marked as unpublished). Sequence comparisions suggest mislabeling is common (11% show best matches to congeneric but heterospecific sequences, and another 7% match among species of a different genus. Overall 10-21% of the INSD sequences have incorrect or unsatisfactory annotations.
The results show a “variegated picture of the taxonomic status of publicly indexed fungal sequences“. Taxonomic coverage is sparse: of the estimated 1.5 million fungi, less than 1% (9,684 species) are represented. Taxonomic data is lacking for many sequences (27% are not identified to species level), and most of the species-level identifications are unverifiable (82% are not linked to voucher specimens, 63% are not tagged with specimen country of origin, and 42% are marked as unpublished). Sequence comparisions suggest mislabeling is common (11% show best matches to congeneric but heterospecific sequences, and another 7% match among species of a different genus. Overall 10-21% of the INSD sequences have incorrect or unsatisfactory annotations.