What is the evidence that DNA barcoding is a reliable method for species identification?
For this commentary, “DNA barcoding” refers to nucleotide sequencing of PCR-amplified DNA corresponding to an approved barcode region, namely 5′ portion of COI for animals or rbcL + matK for land plants; and “species identification” refers to assigning the name of a known species to a specimen of unknown identity.
Acceptance by scientific community. For identification of known species, I think it is fair to say that DNA testing in general and DNA barcoding in particular are generally accepted in the scientific community as reliable methods. For example, the Canadian Centre for DNA Barcoding website has a compilation of peer-reviewed publications, which includes over 500 articles published since 2003. The primary limitation to identification is whether the relevant species and close relatives have yet been documented in the databases at the time they are queried. The BOLD database is strongest for multicellular animals (> 1,000,000 records as of May 2010; see chart), particularly arthropods and chordates. For plants, the general principles are the same, but so far there is much less documentation, as plant barcodes were not agreed-upon until last year (Hollingsworth et al PNAS May 2009), and there was not a large set of pre-existing data to 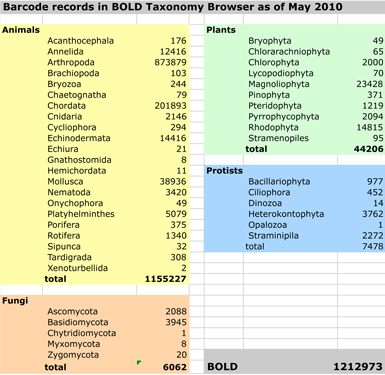 work with. Nonetheless, DNA barcoding of plants is ready for practical application and is providing immediately useful information (e.g. “DNA barcoding exposes a case of mistaken identity in the fern horticultural trade” Prior et al, Mol Ecol Resources April 2010) . For fungi, from perusing database it appears that ITS (internal transcribed spacer) and COI are informally accepted as barcodes. For protists and other domains of life, results so far suggest COI will serve as a primary barcode.
work with. Nonetheless, DNA barcoding of plants is ready for practical application and is providing immediately useful information (e.g. “DNA barcoding exposes a case of mistaken identity in the fern horticultural trade” Prior et al, Mol Ecol Resources April 2010) . For fungi, from perusing database it appears that ITS (internal transcribed spacer) and COI are informally accepted as barcodes. For protists and other domains of life, results so far suggest COI will serve as a primary barcode.
Most articles focus on DNA barcoding in a particular group and assess the accuracy of identification in that group. For example, in “DNA barcoding of commercially important salmon and trout species (Oncorhynchus and Salmo) from North America” (J Agricultural Food Chem 57:8379, 2009) Rasmussen and colleagues analyzed more than 1000 samples representing the 7 commercially important salmonid species from 143 sites across western North America including Alaska and Canada, (to capture possible variation within species) The authors found 100% separation of these species by DNA barcoding, i.e., distances among species were always greater than within species.
Forensic application. DNA barcoding for species identification has been used in legal cases (e.g. Cohen et al J Food Protection 72: 810, 2009). More general evidence is presented by Dawnay et al in “Validation of the barcoding gene COI for use in forensic genetic species identification” (Forensic Sci International 173:1, 2007). The authors conclude “this study demonstrates that the cytochrome c oxidase I gene enables accurate animal species identification where adequate reference sequence data exists.” As with any laboratory method, quality control and quality assurance (QA/QC) measures are essential (e.g. Morin et al J Heredity 101:1, 2010).
DNA barcode identification was designed to be a simple, straightforward method appropriate for wide use, and the results so far amply bear this out, including its use by high school students (e.g., “FDA pressured to combat rising ‘food fraud’,” Lyndsey Layton, Washington Post March 30, 2010). One aspect that needs work in my opinion are better explanations of the algorithms used for matching sequences to the databases and what the results mean. It still takes an expert to make sense of the data. Although the results are often obvious (e.g., 100% sequence identity to 10 barcode records of “Bos taurus (cow)”, interpretation is context dependent–a 100% match has a different meaning if a “neighboring” species differs by, say 1%, or if a congeneric species is not documented or is represented by a single record, for example. In my experience, identifications are usually straightforward, including recognizing ambiguous identifications. Nonetheless, for DNA barcoding to have the widest use, including in legal settings, it will be helpful to have better documentation of how we arrive at species diagnoses through DNA barcodes.
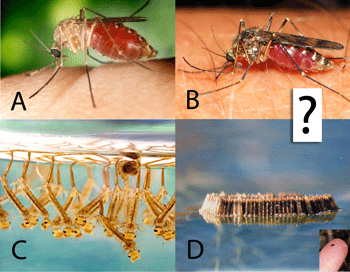 Biting insects transmit human and animal diseases, including protozoan (e.g., malaria, leishmania, trypanosoma (sleeping sickness, Chagas disease)), filiarial (e.g., onchocerciasis, Guinea worm), and viral (e.g., yellow fever, West Nile, dengue) diseases. Control measures rely on identifying the insects, which generally requires expert training.
Biting insects transmit human and animal diseases, including protozoan (e.g., malaria, leishmania, trypanosoma (sleeping sickness, Chagas disease)), filiarial (e.g., onchocerciasis, Guinea worm), and viral (e.g., yellow fever, West Nile, dengue) diseases. Control measures rely on identifying the insects, which generally requires expert training.
 Of the three steps required to get from a specimen to a DNA barcode, namely DNA isolation, PCR (polymerase chain reaction), and sequencing, the first step is the most labor intensive and hardest to automate. Numerous protocols/kits have been developed to optimize DNA isolation from various types of specimens, such as plant vs animal tissues. As described by the Guelph researchers, “these procedures force cells to release their DNA via physical pertubation and/or chemical treatment, which is then followed by a clean-up procedure in which unwanted cellular compoents are separated from the DNA.” The researchers “hypothesized that a small amount of DNA leaks from the tissue into the preservation solution (usually ethanol), and that this DNA was amplifiable using a standard PCR protocol.” To start, they analyzed Monte Alban mescal, which is sold with a “worm” (a caterpillar of the agave moth, Hypopta agavis) in each bottle. They evaporated 50 mL mescal, re-dissolved the residue in water, applied this to a Qiagen MinElute spin column, resuspended the product in 50 ?L water, and used 2 ?L of resulting solution in a standard 25 ?L PCR reaction, with successful amplification and sequencing of 130 base mini-barcode of COI. This case was presumably challenging as mescal is only 40% ethanol and contains a variety of material that might inhibit PCR. In subsequent tests, 1 mL of 95% ethanol used to preserve specimens was evaporated, resuspended in 30 ?L of water without column purification, and 2 ?L used for PCR.
Of the three steps required to get from a specimen to a DNA barcode, namely DNA isolation, PCR (polymerase chain reaction), and sequencing, the first step is the most labor intensive and hardest to automate. Numerous protocols/kits have been developed to optimize DNA isolation from various types of specimens, such as plant vs animal tissues. As described by the Guelph researchers, “these procedures force cells to release their DNA via physical pertubation and/or chemical treatment, which is then followed by a clean-up procedure in which unwanted cellular compoents are separated from the DNA.” The researchers “hypothesized that a small amount of DNA leaks from the tissue into the preservation solution (usually ethanol), and that this DNA was amplifiable using a standard PCR protocol.” To start, they analyzed Monte Alban mescal, which is sold with a “worm” (a caterpillar of the agave moth, Hypopta agavis) in each bottle. They evaporated 50 mL mescal, re-dissolved the residue in water, applied this to a Qiagen MinElute spin column, resuspended the product in 50 ?L water, and used 2 ?L of resulting solution in a standard 25 ?L PCR reaction, with successful amplification and sequencing of 130 base mini-barcode of COI. This case was presumably challenging as mescal is only 40% ethanol and contains a variety of material that might inhibit PCR. In subsequent tests, 1 mL of 95% ethanol used to preserve specimens was evaporated, resuspended in 30 ?L of water without column purification, and 2 ?L used for PCR.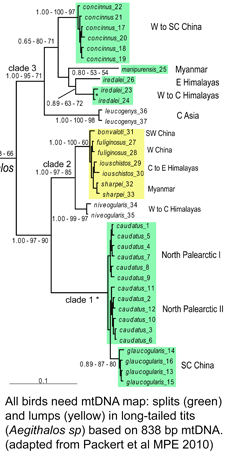 Although birds have been studied in more detail than any other large group of animals, mtDNA continues to reveal many overlooked species, such that named taxa turn out to be comprised of two or more distinct species. These revisions include some very familiar birds, e.g., Canada Goose, which was recently recognized as comprising two species, Cackling Goose (B. hutchinsii) and Canada Goose (B. canadensis) (
Although birds have been studied in more detail than any other large group of animals, mtDNA continues to reveal many overlooked species, such that named taxa turn out to be comprised of two or more distinct species. These revisions include some very familiar birds, e.g., Canada Goose, which was recently recognized as comprising two species, Cackling Goose (B. hutchinsii) and Canada Goose (B. canadensis) (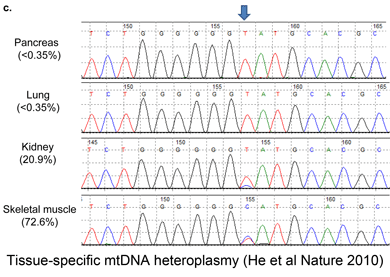 To establish a cutoff for artefactual errors due to PCR and/or sequencing, a control comparison with amplified nuclear DNA was performed, which yielded an average of 0.058% (SD 0.057%) mutations per base and a maximum of 0.82% mutations. He and colleagues used a “very conservative assumption that all variants in excess of twice this value (1.6%) represented true heteroplasmies rather than sequencing artefacts.” Now to some results! The researchers detected “28 homoplasmic alleles and 8 heteroplasmic alleles in this sample of normal colonic mucosa.” Here “homoplastic” refers to differences from the reference human mtDNA sequence (
To establish a cutoff for artefactual errors due to PCR and/or sequencing, a control comparison with amplified nuclear DNA was performed, which yielded an average of 0.058% (SD 0.057%) mutations per base and a maximum of 0.82% mutations. He and colleagues used a “very conservative assumption that all variants in excess of twice this value (1.6%) represented true heteroplasmies rather than sequencing artefacts.” Now to some results! The researchers detected “28 homoplasmic alleles and 8 heteroplasmic alleles in this sample of normal colonic mucosa.” Here “homoplastic” refers to differences from the reference human mtDNA sequence ( In
In  DNA helps answer the origin of infectious diseases: are cases sporadic events or part of larger epidemic, such as the
DNA helps answer the origin of infectious diseases: are cases sporadic events or part of larger epidemic, such as the 
 In terms of citizen participation, the MPG story suggests expanding opportunities for biological research that harnesses the skill and energy of non-professionals, a step beyond the successful
In terms of citizen participation, the MPG story suggests expanding opportunities for biological research that harnesses the skill and energy of non-professionals, a step beyond the successful  Herbal products make a compelling case for DNA-based identification–how else to recognize dried bits of roots, leaves, stems, bark, and flowers from a multitude of species? In
Herbal products make a compelling case for DNA-based identification–how else to recognize dried bits of roots, leaves, stems, bark, and flowers from a multitude of species? In  This study demonstrates advantages of DNA barcoding approach for plant identification. Of course, there is already a lot of interest in DNA identification of herbal plants in general and Dendrobium orchids in particular. For example, I found over a dozen articles describing DNA methods for distinguishing Dendrobium sp. However, the methods described are limited to identifying species in this one genus, which means one has to have a pretty good idea what the specimen is before applying DNA testing! This highlights the essential advantage of barcoding–a standardized approach can be applied to any unknown, and makes feasible creation of a comprehensive reference library.
This study demonstrates advantages of DNA barcoding approach for plant identification. Of course, there is already a lot of interest in DNA identification of herbal plants in general and Dendrobium orchids in particular. For example, I found over a dozen articles describing DNA methods for distinguishing Dendrobium sp. However, the methods described are limited to identifying species in this one genus, which means one has to have a pretty good idea what the specimen is before applying DNA testing! This highlights the essential advantage of barcoding–a standardized approach can be applied to any unknown, and makes feasible creation of a comprehensive reference library.