 Mitochondria are the power plants of the cell, consuming oxygen and breakdown products of sugars, amino acids, and fatty acids to produce energy as ATP and heat. As originally proposed by Lynn Margulis in 1967, mitochondria, which have their own circular genome and replicate independently of the cell, are derived from an ancient symbiosis of an an alpha-proteobacterium related to gram-negative bacteria.
Mitochondria are the power plants of the cell, consuming oxygen and breakdown products of sugars, amino acids, and fatty acids to produce energy as ATP and heat. As originally proposed by Lynn Margulis in 1967, mitochondria, which have their own circular genome and replicate independently of the cell, are derived from an ancient symbiosis of an an alpha-proteobacterium related to gram-negative bacteria.
In multicellular animals, most of the 100+ proteins in mitochondria are encoded by nuclear genes. The mitochondrial genome is only about 16 kb (vs about 2000 kb for nearest bacterial relatives Rickettsia sp) and encodes just 13 proteins, all of which function in the electron transport chain, plus 2 ribosomal RNAs and approximately 20 tRNAs. Multiple protein-protein interactions (for example, complex I comprises 34 nuclear-encoded and 7 mitochondrial-encoded proteins) suggest there must be close co-evolution between nuclear and mitochondrial genomes; this might be one of the constraints on mitochondrial variation. Although an enormous amount of information on mitochondrial sequence differences among and within species has been been compiled (through DNA barcoding initiative and other efforts), there is surprisingly little study so far on whether mitochondrial differences among species reflect functional adaptation (although see Ruiz-Pesini et al 2004 Science 303:223, Bayona-Balfaluy et al 2004 Mol Biol Evol 22:716).
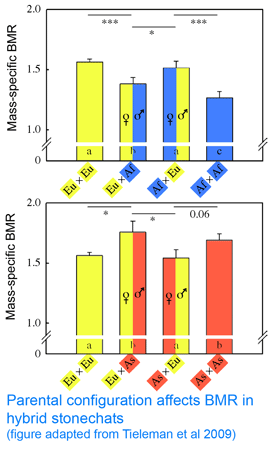 In 25 February 2009 Proc R Soc B researchers from University of Groningen, The Netherlands; Max Planck Institute for Ornithology, Germany; and Ohio State University investigate whether mitochondrial differences modulate energy metabolism in birds. As mitochondria consume 90% of respired oxygen, mitochondrial activity presumably determines basal metabolism. Tieleman and colleagues performed crosses among 3 captive bred populations of stonechats (Saxicola torquata spp.) that differ in basal metabolic rate, which presumably reflects adaptation to different climates: Africa (Kenya, Saxicola torquata axillaris), Asia (Kazakhstan, Saxicola torquata maura), and Europe (Austria, Saxicola torquata rubicola). As an aside, I note that these three taxa are elevated to species status in current world checklists (Clements 2007, IOC Checklist; even 1992 edition of Birds of Europe notes “Siberian race may be a full species.”) This does not change the interpretation of the findings, but it does reflect the confused nature of taxonomic science that even for a group as well studied as birds, publication standards accept this laxity in taxonomic classification. Naming of bacteria in medical studies is more uniformly up to date than for multicellular animals; it seems that animal taxonomists have not found a way to establish a regularly updated consensus. In this regard the IOC Checklist suggests a way forward: “in this global world of wiki-style sharing of knowledge, we invite world birders and ornithologsts alike to help us keep the IOC list accurate, vital, and accessible.”
In 25 February 2009 Proc R Soc B researchers from University of Groningen, The Netherlands; Max Planck Institute for Ornithology, Germany; and Ohio State University investigate whether mitochondrial differences modulate energy metabolism in birds. As mitochondria consume 90% of respired oxygen, mitochondrial activity presumably determines basal metabolism. Tieleman and colleagues performed crosses among 3 captive bred populations of stonechats (Saxicola torquata spp.) that differ in basal metabolic rate, which presumably reflects adaptation to different climates: Africa (Kenya, Saxicola torquata axillaris), Asia (Kazakhstan, Saxicola torquata maura), and Europe (Austria, Saxicola torquata rubicola). As an aside, I note that these three taxa are elevated to species status in current world checklists (Clements 2007, IOC Checklist; even 1992 edition of Birds of Europe notes “Siberian race may be a full species.”) This does not change the interpretation of the findings, but it does reflect the confused nature of taxonomic science that even for a group as well studied as birds, publication standards accept this laxity in taxonomic classification. Naming of bacteria in medical studies is more uniformly up to date than for multicellular animals; it seems that animal taxonomists have not found a way to establish a regularly updated consensus. In this regard the IOC Checklist suggests a way forward: “in this global world of wiki-style sharing of knowledge, we invite world birders and ornithologsts alike to help us keep the IOC list accurate, vital, and accessible.”
Back to the paper. Tieleman and colleagues “tested for a genetic effect on BMR based on mitochondrial-nuclear coadaptation using hybrids between ancestral populations with high and low BMR (Europe-Africa and Asia-Europe), with different parental configurations (female-high x male-low or female-low x male-high). Hybrids with different parental configurations have on average identical mixtures of nuclear DNA, but differ in mitochondrial DNA because it is inherited only from the mother.” The researchers found that metabolic rate differed between hybrids with contrasting parental configurations, “providing evidence for the importance of a match between mitochondrial and nuclear genomes to regulate metabolic rate.” So far so good. However, contrary to expectations, in both sets of crosses, metabolic rates in hybrids were more similar to that of the father than the mother! (see adapted figure). This result is a puzzler; it suggests there might be another factor such as genomic imprinting at work.
Looking at the bigger picture, for those interested in mitochondrial evolution, there is a lot of opportunity: a large and growing database of COI sequences (>500,000 individuals, >50,000 species so far) that is waiting to be analyzed for evidence of purifying or positive selection, for example, or for limits to plasticity in COI amino acid sequence. I wonder if there might be convergent evolution of COI, such that diverse organisms in very cold or very hot environments environments, for example, might exhibit similar amino acid substitutions.