What do leaf beetles (Chrysomelidae) eat? In 11 Nov 2008 Proc R Soc B researchers from Spain, London, and Australia, apply DNA analysis to 76 species (1 individual/species) of Australian leaf beetles. Jurado-Rivera and colleagues extracted DNA from whole beetles using DNAeasy kit. To identify plant DNA in beetle extracts, they amplified chloroplast trnL intron (313 to 581 bp in analyzed samples). 70 (92%) of samples gave high quality reads after direct sequencing of the PCR products, consistent with ingestion of a single plant species; the remaining samples were sequenced from cloned PCR products; these gave 2 divergent clones in 3 of seven cases, for a total of 81 different trnL intron sequences. Why use trnL intron? The authors cite the large number of sequences in GenBank and favorable experience (ie successful amplification and good taxonomic resolution) in their prior work and that of others (eg Taberlet et al 2007 Nucleic Acids Res 35:e14). This certainly makes sense, but I hope a general agreement for plant barcode standards will be published shortly, otherwise the field will continue to be hobbled by having multiple incomplete and non-overlapping databases for the various markers. For example, according to the authors “only 14 and 15 of approximately 1000 and 800 described Australian species of Acacia and Eucalyptus, respectively, are represented in GenBank by trnL intron sequences. As to what resolution is possible with current trnL database, the authors found “reliable identification to plant family in every case and very frequently the inference is possible at lower taxonomic levels.”
What do leaf beetles (Chrysomelidae) eat? In 11 Nov 2008 Proc R Soc B researchers from Spain, London, and Australia, apply DNA analysis to 76 species (1 individual/species) of Australian leaf beetles. Jurado-Rivera and colleagues extracted DNA from whole beetles using DNAeasy kit. To identify plant DNA in beetle extracts, they amplified chloroplast trnL intron (313 to 581 bp in analyzed samples). 70 (92%) of samples gave high quality reads after direct sequencing of the PCR products, consistent with ingestion of a single plant species; the remaining samples were sequenced from cloned PCR products; these gave 2 divergent clones in 3 of seven cases, for a total of 81 different trnL intron sequences. Why use trnL intron? The authors cite the large number of sequences in GenBank and favorable experience (ie successful amplification and good taxonomic resolution) in their prior work and that of others (eg Taberlet et al 2007 Nucleic Acids Res 35:e14). This certainly makes sense, but I hope a general agreement for plant barcode standards will be published shortly, otherwise the field will continue to be hobbled by having multiple incomplete and non-overlapping databases for the various markers. For example, according to the authors “only 14 and 15 of approximately 1000 and 800 described Australian species of Acacia and Eucalyptus, respectively, are represented in GenBank by trnL intron sequences. As to what resolution is possible with current trnL database, the authors found “reliable identification to plant family in every case and very frequently the inference is possible at lower taxonomic levels.”
There also needs to be an agreement to have a curated plant barcode database. As the authors report, “in the course of this study, we found several examples of erroneous taxonomic assignments (e.g. Sapindaceae identified as Cypripedium, Cypripedioideae; Apocynaceae labelled as Sesamum, Pedaliaceae; one case of names switched between Pittosporum and Cheiranthera, both Pittosporaceae; suspicious generic assignment for Aesculus x carnea), and of sequencing artefacts (e.g. Tragopogon spp., Acacia usumatensis) and chimeras (e.g.Pentaphylax euryoides). Problems introduced by these sequences were only apparent after careful inspection of trees revealing suspicious relationships, and required phylogenetic re-evaluation after removing problematic sequence data.”
This is helpful for the present study, but the problematic sequences remain in the reference databases, ready to trip up the next set of researchers who might not be so careful. To fix this problem, Jurado-Rivera and colleagues make what I think is the wrong suggestion, namely “all of the above would argue for the use of additional markers”. Adding markers may improve the ability to make species-level identifications in plants, but if the goal is to construct an error-free database, adding markers is an expensive and likely ineffective way to ferret out mislabeled or otherwise inaccurate sequences. What is needed is a stand-alone database, closely-linked to GenBank, in which problematic sequences can be weeded out or re-labeled (ie Barcode of Life Database (BOLD) www.barcodinglife.org).
To construct a beetle phylogeny, the authors amplified COI and EF1a from their specimens. They found strong concordance between the evolutionary histories of Australian Chrysomelinae beetles and their host plants, indicating long-term co-evolution. They conclude “our analysis not only shows the details of ecological associations for a dominant herbivore group but also offers the basis for their evolutionary interpretation.”
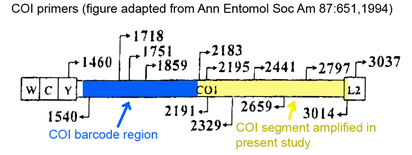
I am puzzled that the authors amplified a segment in the 3′ half of COI that does not overlap with the standard animal barcode region, making it impossible to combine their data with the 500,000+ COI sequences analyzed to date (www.barcodinglife.org). This important caveat aside, I look forward to many more studies that utilize DNA barcoding to join ecology and phylogenetics.