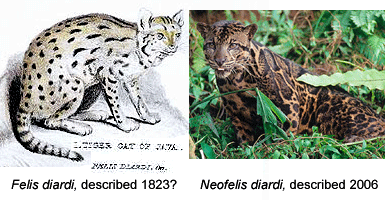
Like a telescope that reveals hidden structures in the universe, genomic analysis is a window into biodiversity. For one, differences in DNA sequences help reveal how biodiversity is partitioned into the distinct populations we call species. In Frontiers Zool 29 May 2007, researchers from University of Wurzburg, US National Cancer Institute, and Arizona State University report on mitochondrial DNA and nuclear microsatellite differences between clouded leopards (Neofelis nebulosa) from Borneo (5 individuals), Sumatra (2 individuals), and mainland SE Asia (6 individuals). This report is a follow-up on two papers in December 2006 Current Biol which proposed separate species status for Bornean clouded leopards on the basis of differences in coat pattern and DNA. Wilting et al conclude their updated results “strongly support reclassification of clouded leopards into two distinct species N. nebulosa and N. diardi“. In addition to distinct coat patterns, the two lineages differ by 4.5% in mitochondrial coding genes (cytochrome b and ATPase-8), equivalent to or larger than genetic distances between the other well-recognized species of big cats in Panthera genus (lion, jaguar, tiger, leopard, snow leopard), suggesting the two lineages of clouded leopards have been separated for about 2.86 million years.
This sounds straightforward, but some taxonomists lament the increasing role of DNA in species discovery. In an editorial in current PLoS ONE, researchers from Imperial College insist the Bornean clouded leopard is not really new as it was “described by Cuvier in 1823.” Of course, by this criteria, most forms of larger animals will have been “described” by someone. Cuvier’s original work naming Felis diardi is three short paragraphs based on a single specimen and the illustration is unrecognizable.
To my reading, Meiri and Mace’s editorial implies that most of the important taxonomic work has already been done and if new genetic data appear to upset the traditional scheme, then it is being incorrectly interpreted. They note that there are another 144 mammal species shared between Borneo and the Malay Peninsula, thus “there could potentially be equivalent evidence to merit specific status for all of these; an outcome that would surely be unjustified”. An outcome that would surely be unjustified? This question needs to be answered by science, not by an appeal to taxonomic tradition. It may be that many island populations, which are now considered allopatric forms of widely distributed species, will turn out to be distinct species.
I close with the observation that just as genetic data can suggest splits it can also help reveal synonomies (multiple names that refer to the same species), suggest lumps, and identify forms that do NOT merit separate conservation status. For example, in Proc R Soc B 2005 Johnson et al apply mitochondrial DNA analysis to argue that the Cape Verde kite is not genetically distinct from the Black kite Milvus migrans and does not merit separate conservation status.

There is a clear disconnect between trees of splitting mitochondrial non-coding loci and trees of evolution of expressed traits involved in actual speciation. The genetic distance merely means that the two populations have been isolated a long time, but apparently expressed traits indicate that balancing evolution (or some such) has kept the two populations identical in morphology and probably evolutionary ecology. No speciation.
What has really changed is the definition of evolution as hereditable changes in non-codong loci. Just a couple decades ago, genetics had to do with expressed traits, then non-coding bases were used to approximately track changes in expressed traits (via biological species concept), then genetics as far as systematics is concerned involves only non-coding loci.
If the DNA bases are coding, then selection can be involved, and convergence, theoretically avoided with non-coding DNA bases, confounds molecular trees and further disconnects them from speciation involving expressed traits (e.g. selection or drift). What has happened is that spandrels as expressed traits have been conflated with non-coding base changes, and if we focus entirely on non-coding bases, we neglect entirely speciation by selection.