In Barcoding Life, Illustrated we suggested that “standardization typically lowers costs and lifts reliability…and should help accelerate construction of comprehensive, consistent reference library of DNA sequences and development of economical technologies for species identification.” However, some have been doubtful about the benefits of standardization. This reflects in part the balkanized nature of taxonomic science, as most systematists specialize on groups of related organisms and have limited scientific interactions with those studying other groups. Last year Jesse Ausubel and Paul Waggoner outlined some of the concerns raised by the growth of DNA barcoding in Barcoding Worries and Limits. The final item asks whether “it is too soon to standardize on a very few localities”. The salamander paper discussed in last week’s post seems to support this concern, as it implies that the relative rates of evolution of mitochondrial genes differ between animal groups. According to the paper, in salamanders COI evolves much more rapidly than other mitochondrial protein-coding genes.
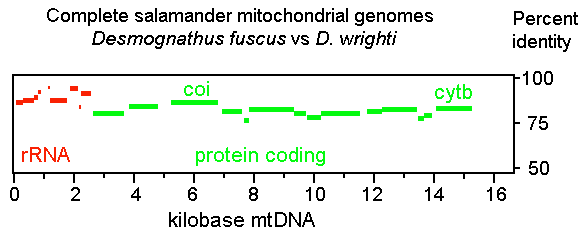
However, as detailed below, my analysis indicates that salamanders are unexceptional in terms of COI differences. I find evolutionary divergences are widely distributed in mitochondrial protein-coding genes and the patterning of these differences is similar in diverse invertebrate and vertebrate organisms, including salamanders. First, I used PipMaker (percent identity plot) to examine the distribution of sequence differences in complete mitochondrial genomes. A representative PipMaker plot, shown above, compares the mitochondrial genomes of 2 plethodontid salamanders, Desmognathus fuscus and D. wrighti, revealing a typical pattern of widely distributed sequence differences. As in most vertebrates, in salamanders COI is slightly LESS divergent (14%) than other protein-coding regions, including cytochrome b (CYTB) (17%).
Second, I compiled differences between closely-related species pairs in CYTB and COI. For historical reasons, CYTB has been the single most common locus used to analyze differences in vertebrate species, and COI has been the single most common single locus for invertebrates. As shown below, these genes show highly correlated differences in deeply divergent lineages of invertebrates and vertebrates, including salamanders.
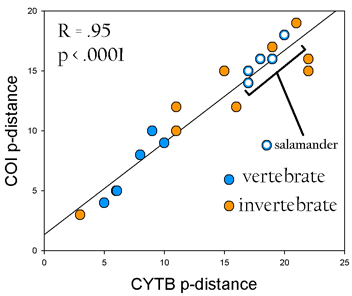
Species examined include Demospongiae (sponges); Platyhelminthes (tapeworms); Echinodermata (starfish); Arthropoda (ticks; fruit flies; mosquitos; shrimp); Mollusca (octopi; mantis shrimp) Vertebrata (turtles; eels; coelecanths; elephants; wolves; apes; and 5 pairs of congeneric salamanders)
These findings support standardizing on COI as the primary DNA barcode for multicellular animals. As discussed in earlier posts, there will of course be cases in which the primary barcode does not resolve closely-related species and a secondary barcode(s) may be needed.