Sushigate continues! The discovery of inaccurately labeled fish by Kate Stoeckle and Louisa Strauss (see 22 August 2008 What’s New entry) evoked a NY Times sequel story where chefs claimed supreme expertise, but then today Edward Dolnick countered with a wise Op-Ed “Fish or Foul” about the judgment of experts.
News
Dominique de Saint Pern
The French newspaper Le Monde on 15 August 2008 featured an excellent article
and an interview by Dominique de Saint Pern about the technical and scientific
progress of the film about Oceans by Jacques Perrin and Galatee films, which
Jesse and the Census of Marine Life have had the privilege of assisting. We
post English translations by Vesna Neskow of the article and the
interview as well as the original French article and interview.
CSI for fish: High school students help showcase ease of DNA-based species identification
 In Pacific Fishing September 2008 (issue available on newstands, not yet on web) two New York City teenagers, Kate Stoeckle (my daughter!), 19, and classmate Louisa Strauss, 18, apply DNA-based identification to fish sold in their Manhattan neighborhood. The girls purchased 60 items from 14 establishments and sent samples to University of Guelph where graduate student Eugene Wong performed DNA barcode analysis. 14 (25%) of 56 samples with recoverable DNA were mislabeled, in all cases as more expensive or more desirable fish. Mislabeled items were sold at 2 of 4 restaurants and 6 of 10 grocery stores/fish markets.
In Pacific Fishing September 2008 (issue available on newstands, not yet on web) two New York City teenagers, Kate Stoeckle (my daughter!), 19, and classmate Louisa Strauss, 18, apply DNA-based identification to fish sold in their Manhattan neighborhood. The girls purchased 60 items from 14 establishments and sent samples to University of Guelph where graduate student Eugene Wong performed DNA barcode analysis. 14 (25%) of 56 samples with recoverable DNA were mislabeled, in all cases as more expensive or more desirable fish. Mislabeled items were sold at 2 of 4 restaurants and 6 of 10 grocery stores/fish markets.
The frequency of mislabeling and the ease of high-schoolers obtaining DNA-based species identifications captured public interest. Their study was featured on page 1 in New York Times on August 22, one of the “quotes of the day” on CNN/Time website, a live segment on CBS TV Early Show, an interview on national public radio, and has appeared in over 350 print and news items and blogs from 34 countries in 10 languages so far, with particularly heavy coverage in China, Korea, and Japan, presumably related to dietary importance of fish in general and sushi in particular.
This response demonstrates how powerful the FishBOL database is already (30,665 barcodes from 5,463 species so far, which represents about 20% of world fish), and hints at enormous uses that DNA barcoding will have as technology gets smaller and cheaper.
PNAS Dematerialization
For generations, people have lightened their environmental impact
by multiplying their consumption less than their income. A
combination of consumers tempering their consumption of goods and
producers making the goods with less harm to the environment has
long moderated human impact. Does recent experience justify hope
for sustaining this beneficial dematerialization, especially the
decarbonization of national and global carbon emission?
The Proceedings of the National Academy of Sciences (PNAS) has
just published an answer, “Dematerialization: Variety, Cautionand Persistence” by Rockefeller’s Jesse Ausubel and
Paul Waggoner of the Connecticut Agricultural Experiment Station.
The PNAS report addresses whether the impact of consumers and
producers is growing heavier or lighter and finds a variety of
hopeful examples from energy and carbon emission through wood
consumption on to farming and land use. The report unfortunately
also finds discrepancies and fluctuations in data that require
caution in drawing generalizations and that need remedying to
avoid missteps.
Nevertheless, encouraging evidence, especially for
dematerialization by consumers, prevails. While the paper
reports troubling directions for Brazil and especially Indonesia,
India by several reports changed from a worsening to an improving
environmental performance. Chinese dematerialization slowed a
bit, but did not cease by any report, and the rise of its
intensity of impact either slowed or reversed. Surprisingly,
apparently unaffected by changes of government, the U.S.A.
dematerialized steadily near 2%/yr throughout the period 1980-2004.
Fish DNA Barcode CBS morning show
Kate and Mark Stoeckle appeared on the CBS Morning Show
discussing “Shocking Sushi Secrets” based on Kate and Louisa
Straus’ study of fish identity using DNA barcoding. The 4-minute
video is at:
https://www.cbsnews.com/video/watch/?id=4377085n%253fsource=search_video
Students Use DNA Barcodes to Unmask “Mislabeled” Fish at Grocery Stores, Restaurants
Pacific Fishing magazine has published the report on mislabeled fish identified by DNA barcodes by star students Kate Stoeckle and Louisa Strauss, whom PHE had the pleasure of assisting. For their report, pictures of some of the fish, and related information below. Their work also earned front-page coverage in The New York Times.
- High school friends make first student use of DNA barcodes in public marketplaces
- One-quarter of 56 fish samples from 14 stores, restaurants in Upper Manhattan revealed to be cheaper or endangered fish species.
- Mozambique Tilapia sold as “White Tuna” in sushi
Two New York City high school friends, curious about new DNA barcoding technology, discovered that fish at local stores and restaurants are commonly mislabeled and sold for far more than regular market price.
Worse, in two cases DNA barcode tests revealed that filleted fish sold as the popular Red Snapper (caught mostly off the southeast U.S. and in the Caribbean) was instead the endangered Acadian Redfish (which swims in the North Atlantic).
The students’ report marks the first marketplace application of the four-year-old DNA barcoding technology.
Contact
Mr. Terry Collins
416-538-8712
terrycollins@rogers.com
Jesse Ausubel
Program for the Human Environment, The Rockefeller University
212-327-7842
ausubel@rockefeller.edu
Kevin Ramsey
Trinity School
kevin.ramsey@trinityschoolnyc.org
Documents
a related paper on identifying Canadian freshwater fishes through DNA barcoding
Images

Albacore Tuna Thunnus alalunga
Mozambique Tilapia Oreochromis mossambicus

Red Snapper Lutjanus campechanus
Lavender Jobfish Pristipomoides sieboldii
Slender Pinjalo Pinjalo lewisi
Nile Perch Lates niloticus
Acadian Redfish Sebastes fasciatus
The authors: Kate Stoeckle and Louisa Strauss
Useful links
- Encyclopedia of Life: www.eol.org
- Barcode of Life Database: www.barcodinglife.org
- Consortium for the Barcode of Life: barcoding.si.edu
- Barcoding marine species: www.marinebarcoding.org
- FishBol: www.fishbol.org
- Barcoding blog: https://phe.rockefeller.edu/barcode/blog
- Ten Reasons for Barcoding Life: https://phe.rockefeller.edu/barcode/docs/TenReasonsBarcoding.pdf
- “Barcode of Life” Scientific American, October 2008: https://phe.rockefeller.edu/docs/BarcodeScientificAmerican.2008.10.pdf
Decarbonization in The American
Energy author Robert Bryce publishes a piece in The American
magazine that refers heavily to our work on Decarbonization.
Summer hiatus
I will be away from Blog until mid-August.

Helping reveal relationships among species
COI barcodes aim to enable identification of species, assigning unknown specimens to known species, and helping flag genetically divergent organisms that may represent new species. Might barcodes also help understand relationships among species?
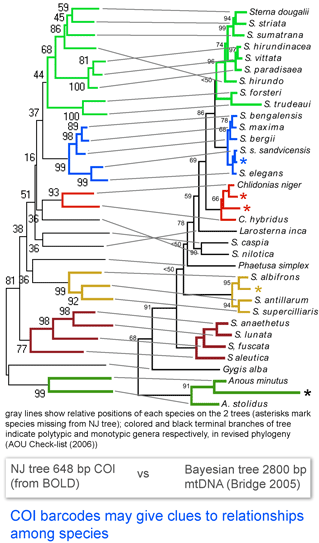 Here I look at one example from birds, comparing differences among COI barcodes to a recently revised phylogeny of terns (subfamily Sternini). According to American Ornithologists’ Union Check-list of North American Birds Supplement 47 (2006), “the data show that the genus Sterna as currently defined…is paraphyletic.”…”[W]e follow the recommendation of Bridge et al 2005 to resurrect four generic names currently placed in synonomy with Sterna.” The figure at left, taken from the 2005 paper by Bridge, Jones, and Baker, shows phylogenetic relationships based on 2800 bp of mtDNA from 33 species of terns (Bayesian tree with ML distances and ML boostrap support indices), and is juxtaposed to an NJ tree of COI barcodes from 29 of the same 33 taxa. The figure is colored according to the revised generic assignments (AOU 2006).
Here I look at one example from birds, comparing differences among COI barcodes to a recently revised phylogeny of terns (subfamily Sternini). According to American Ornithologists’ Union Check-list of North American Birds Supplement 47 (2006), “the data show that the genus Sterna as currently defined…is paraphyletic.”…”[W]e follow the recommendation of Bridge et al 2005 to resurrect four generic names currently placed in synonomy with Sterna.” The figure at left, taken from the 2005 paper by Bridge, Jones, and Baker, shows phylogenetic relationships based on 2800 bp of mtDNA from 33 species of terns (Bayesian tree with ML distances and ML boostrap support indices), and is juxtaposed to an NJ tree of COI barcodes from 29 of the same 33 taxa. The figure is colored according to the revised generic assignments (AOU 2006).
The topology of the COI NJ tree is similar to the larger data set tree, including that all currently recognized genera are reciprocally monophyletic, and most show similarly high boostrap values as in the Bayesian/ML analysis based on the larger data set.
Of course mitochondrial DNA is widely used in analyzing relationships among animal species, including birds. Most of these studies are focused on relatively small groups of species, such as the tern study cited here. With growing DNA barcode libraries it will be increasingly possible to get at least a preliminary look at genetic relationships for large numbers of species (so far 2,393 avian species (24% of world birds) have barcode records in BOLD). This could be exciting!
Mitochondrial DNA’s unique power
The DNA barcode for animals is a 648 base pair (bp) fragment from the 5′ end of mitochondrial gene cytochrome c oxidase subunit I (COI). Does this relatively short mitochondrial sequence contain enough information to make evolutionary inferences about species limits, or is it a more of a rough survey method that needs to be confirmed by more data including from nuclear genes?
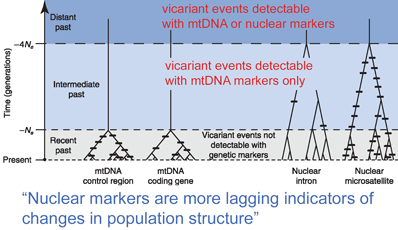 In April 2008 Mol Ecol researchers from University of Minnesota and American Museum of Natural History, New York, analyze utility of mitochondrial as compared to nuclear DNA for inferring recent evolutionary history. Zink and Barrowclough first apply population genetic theory and then look at real data from bird species.
In April 2008 Mol Ecol researchers from University of Minnesota and American Museum of Natural History, New York, analyze utility of mitochondrial as compared to nuclear DNA for inferring recent evolutionary history. Zink and Barrowclough first apply population genetic theory and then look at real data from bird species.
Based on mathematical population genetics, they find “mitochondrial loci are generally a more sensitive indicator of population structure than are nuclear loci,” primarily due to much smaller effective population size (Ne) for mitochondrial as compared to nuclear markers, which leads to more rapid sorting of differences among genetically isolated populations. Analysis of real-world data in 45 studies of differences among and within avian species confirms this expectation, ie either the patterning is consistent between mitochondrial and nuclear genes, or there are shallow mtDNA trees which are not yet reflected in nuclear genes. Reanalysis of one study, which appears to show a split in nuclear but not mitochondrial markers, suggests possible misinterpretation. Regarding other factors that could potentially lead to mistaken inferences about species limits based on mitochondrial DNA (NUMTs, sex-biased gene flow, introgression), experimental data suggests these are rarely important, at least in birds. The authors conclude “mtDNA patterns will prove to be robust indicators of population history and species limits.” Nuclear markers ARE important for deep gene trees, for detecting hybrids, and for “quantitative estimates…of rates of population growth and values of gene flow.”
Regarding length of mitochondrial sequence, this only has to be long enough to capture differences among closely-related species. Most populations that we recognize as species differ from their closest relatives by >1% in mitochondrial coding regions (corresponding to about 0.5 million years or more of reproductive isolation). At this level, even 100 bp is generally sufficient to distinguish most closely-related species, and a 648 bp COI barcode sequence should generally allow resolution of populations/species which have been reproductively isolated for much shorter periods of time.

