Mitochondria are energy-producing organelles, found in nearly every cell in nearly every plant and animal species (some protozoans lack mitochondria). As first demonstrated by Lynn Margulis in 1967 (J Theor Biol 14:225) mitochondria, like chloroplasts, are derived from bacteria, reflecting an ancient symbosis that pre-dates the divergence of plant and animal species 1 billion years ago. Both organelles have retained a truncated circular genome and replicate independently of the nucleus. The mitochondrial genome in particular has turned out to be exceedingly useful in tracing evolutionary history, as it is present in all eukaryotic organisms, evolves rapidly as compared to nuclear DNA, and does not undergo meiosis and recombination, processes that scramble the evolutionary lineages of nuclear genes. Because it is several orders of magnitude more abundant than nuclear DNA (hundreds to thousands of mitochondria per cell, and 5 to 10 genomes per mitochondrion), it facilitates forensic identification, including DNA barcoding, even with very small or degraded samples. Given its practical and scientific importance, we want to better understand mitochondrial genetics.
A general observation is that mitochondrial DNA is uniform in an individual, presumably reflecting a stringent bottleneck at oogenesis, such that one or a very small number of mitochondria are passed on. However, a number of cases of heteroplasmy (i.e., individuals harboring multiple mitochondrial variants) are reported in humans and other animals. In 3 March 2010 Nature researchers from Johns Hopkins University and Case Western University apply next-generation sequencing to make a high-resolution survey of mitochondrial heteroplasmy in various tissues including cancerous cells. He and colleagues used “two sets of PCR primers, each resulting in amplicons of about 650 base pairs (bp) in length…to cover the mtDNA genome” and the resulting PCR products were sequenced by synthesis in an Illumina GAII. This approach was applied to a sample of normal colonic mucosa, yielding “8.5 million tags that matched the mitochondrial genome.” As a result, “each mtDNA base was sequenced, on average 16,700 times and fewer than 11 bases (0.07%) of the 16,569 bp in the mtDNA genome) were represented fewer than 1,000 times.”
 To establish a cutoff for artefactual errors due to PCR and/or sequencing, a control comparison with amplified nuclear DNA was performed, which yielded an average of 0.058% (SD 0.057%) mutations per base and a maximum of 0.82% mutations. He and colleagues used a “very conservative assumption that all variants in excess of twice this value (1.6%) represented true heteroplasmies rather than sequencing artefacts.” Now to some results! The researchers detected “28 homoplasmic alleles and 8 heteroplasmic alleles in this sample of normal colonic mucosa.” Here “homoplastic” refers to differences from the reference human mtDNA sequence (NCBI entry NC_012920). All of the homoplastic alleles were previously found in normal individuals, so we can set these aside as representing normal variation among human individuals.
To establish a cutoff for artefactual errors due to PCR and/or sequencing, a control comparison with amplified nuclear DNA was performed, which yielded an average of 0.058% (SD 0.057%) mutations per base and a maximum of 0.82% mutations. He and colleagues used a “very conservative assumption that all variants in excess of twice this value (1.6%) represented true heteroplasmies rather than sequencing artefacts.” Now to some results! The researchers detected “28 homoplasmic alleles and 8 heteroplasmic alleles in this sample of normal colonic mucosa.” Here “homoplastic” refers to differences from the reference human mtDNA sequence (NCBI entry NC_012920). All of the homoplastic alleles were previously found in normal individuals, so we can set these aside as representing normal variation among human individuals.
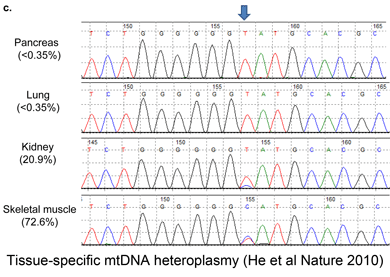
The researchers extended this analysis to other tissues from same individual; all tissues yielded heteroplasmic mtDNA, and the proportion of of individual variants differed strongly among tissues, e.g., the frequency of the most common variant ranged from 7.4% in skeletal muscle to 90.9% in kidney. Surprisingly, 75% of the heteroplasmic variants are already reported in human databases, suggesting a limited pool of variation and/or strong purifying selection. Further evidence for restricted variation is that 67% of heteroplasmic variants were in non-protein coding or RNA-coding regions,” presumably the control region, which represents less than 10% of the mitochondrial genome.
What is the origin of heteroplasmic mtDNA? Using samples from one kindred, the researchers found identical variants in a mother and her two children (and not in the father), demonstrating that, at least in this case, the heteroplasmic variants were inherited from the mother. The authors go on to analyze mitochondrial heteroplasmy in cancerous tissue which is interesting but I will not discuss here.
In terms of species-level identification, the findings add confidence to the established approach using COI mtDNA for animals. This high-resolution study demonstrates that mitochondrial variation within human individuals is a smaller scale version of the variation already known to exist among individuals. As in standard mitochondrial genetics, most of the heteroplasmic variants are maternally inherited. On the other hand, when identification of individuals is important, as in human forensics, mitochondrial heteroplasmy may need to be taken in account, at least on the negative side when apparent mismatches are found. The authors conclude by suggesting “caution in excluding identity on the basis of a single or small number of mismatched alleles when the tissue in evidence (such as sperm) is not the same as the reference tissue of the suspect (such as blood or hair).” Looking ahead, for those interested in exploring mitochondrial heteroplasmy in other species, the initiative has created a large database of intra-specific variation in diverse species, an essential benchmark for investigating possible within-individual variation.
Note added 22 march 2010: As a thought experiment we can ask: how much of the within-species variability in COI might be due to unrecognized mitochondrial heteroplasmy? In the present study, the average number of heteroplasmic variant sites in one tissue sample was about 5, and, on average, 33% of such sites were in protein- or RNA-coding regions, which represent about 90% of mitochondrial genome. That gives (5 x .33) sites distributed across (16,569 x 0.90) nucleotides in the mitochondrial genome, which works out to about 0.0001 variants per site. For 650 bp COI barcode region, that corresponds to an average of 0.07 heteroplasmic sites per barcode sequence, or 0.01% variation. So for humans at least, mitochondrial heteroplasmy appears unlikely to contribute significantly to the observed intra-specific variation in COI.