How does one collect tropical reef fish without leaving North America? In July 2009 PLos ONE researchers from University of Guelph report on genetic diversity in SE Asian tropical reef fish, collected without plane fares or permits. How did they do it? Steinke and colleagues analyzed “dead on arrival” marine fish imported into Canada for the ornamental pet trade from various locations in SE Asia. A total of 1631 specimens representing 391 named species were frozen, imaged on a flatbed scanner, and a muscle tissue sample was taken for COI analysis. This is remarkable on several counts. First, the large number of species–according to FAO report cited by Steinke, “some 800 marine fish species, representing about 5% of all marine taxa, are involved in this trade, with 70% of sales directed to North America,” and estimated revenue of $200-$300 million annually. Second, this study surveys genetic biodiversity in reef fishes, provides a practical method for identification, and at the same time provides insight into what is probably the major threat to their survival. I am reminded of near extinction of Common egrets in North America in the late 1800’s as a result of hunting for plumes in women’s hats. This led to a popular uprising among women of fashion, who pledged not to wear such clothing, organizing what were the first “Audubon Societies” and successfully petitioning for legislative change, saving egrets and many other birds. Nemo and other reef fish may need a similar campaign.
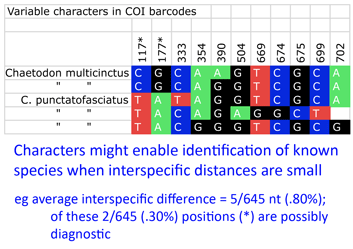 Back to the study, Steinke and colleagues found distinct barcodes among 384/391 (98.2%); 9 species displayed 2 or 3 distinct clusters, most of which were allopatric. Review of these potential “splits” revealed possible inappropriate synonymization in several cases. On the other side, 2 pairs and 1 triplet of species were not distinguished by DNA barcodes using distance. I look more closely at one of these examples, butterfly fishes Chaetodon multicinctus and C. punctatofasciatus, to see if there might be diagnostic characters whose signal is swamped by intraspecific variation. As in figure, there are 2 possibly diagnostic differences among this species pair. Of course, this sort of analysis only works for known species, but I wonder how many other species pairs/sets with “overlapping” barcodes have diagnostic differences.
Back to the study, Steinke and colleagues found distinct barcodes among 384/391 (98.2%); 9 species displayed 2 or 3 distinct clusters, most of which were allopatric. Review of these potential “splits” revealed possible inappropriate synonymization in several cases. On the other side, 2 pairs and 1 triplet of species were not distinguished by DNA barcodes using distance. I look more closely at one of these examples, butterfly fishes Chaetodon multicinctus and C. punctatofasciatus, to see if there might be diagnostic characters whose signal is swamped by intraspecific variation. As in figure, there are 2 possibly diagnostic differences among this species pair. Of course, this sort of analysis only works for known species, but I wonder how many other species pairs/sets with “overlapping” barcodes have diagnostic differences.
This is more good work by the barcoding team up in Guelph. I’d like to make an observation or two – unfortunately, these sequences don’t seem to be available on GenBank yet…preventing one from checking if these are “Barcode” keyworded specimens – we too have gathered data on hundreds of species from the aquarium trade, but gathering factually correct collection localities seems near impossible – the importer doesn’t want to give up their supplier, the supplier doesn’t want to reveal their reef-fish collecting localities, and nobody wants to get caught in areas they are not supposed to be collecting in (or using methods they are not supposed to – remember they are live collecting and these things ended up dying)- this leads to difficulty establishing the true provenance of the specimens which in turn leads to difficulty in interpreting the possible causes of differentiation (closely related species or big geographical variation?). In their BOLD project, it indicates that most have geo-referencing, but with what degree of certainty? While grabbing these imports is easy and efficient, we get much more reliable data by scientifically collecting our specimens ourselves. Our data indicate that an ocean basin or a country’s waters is not precise enough. I don’t disagree with much in this, just that the weaknesses of the approach weren’t even mentioned. I’d hate to encourage this as the standard to which new fish barcoders should strive – it is a compromise. Our fish group wouldn’t rank the aquarium trade in the top two reasons threatening global reef fish – those being global warming and habitat loss/degradation – if we could stop collecting today but if they don’t have a healthy place to live and reproduce, that effort would be for naught. Many of the species also become the subjects of captive breeding research – Nemo, and all the clown fish species being a prime example. The captive bred tend to be easier to keep and maintain, as well as less expensive – a win-win for all.