Phylogenetic tree-building programs are the workhorses of evolutionary analysis. Thus it might be surprising that, given there are at least 1.7 million named species of plants and animals, output trees with over 1000 taxa are exceptional. The primary reason is computational–the number of possible arrangements rises logarithmically with input taxa (eg for 1000 taxa, ~10^2500 possible trees; Tamura et al 2004), such that standard algorithms, even those that sample a fraction of “tree space,” are too slow. As a result, so far the Tree of Life has been constructed by concatenating multitudes of trees each built with relatively small numbers of taxa. This is unsatisfying and possibly unreliable.
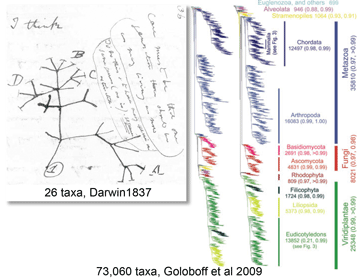 In May 2009 Cladistics researchers from Argentina and Sweden report on the largest tree to date–73,060 eukaryotic taxa, essentially everything Goloboff and colleagues could find in GenBank, ranging from algae and protozoans to flowering plants and vertebrates. In addition to size, there were several remarkable features. The tree was constructed from just 13 genes, each of which was sequenced for a subset of the total (750 to ~20,000 taxa), plus 604 morphologic characters that applied across most of the data set. Nearly all (92%) of the cells in the resulting data matrix (73,060 taxa x 9535 characters) were empty due to lack of data. Nonetheless, the parsimony analysis recovered most eukaryotic groups down to the level of order as monophyletic taxa. The analysis utilized TNT software previously developed (and made publicly available) by Goloboff and colleagues and took 2.5 months on 3 desktop computers (total 96 GB RAM, 16 x 3 Ghz processors). To manage the flow of data, nearly all steps were automated from extracting, labeling, and aligning GenBank sequences to analyzing monophyly of groups at various taxonomic levels.
In May 2009 Cladistics researchers from Argentina and Sweden report on the largest tree to date–73,060 eukaryotic taxa, essentially everything Goloboff and colleagues could find in GenBank, ranging from algae and protozoans to flowering plants and vertebrates. In addition to size, there were several remarkable features. The tree was constructed from just 13 genes, each of which was sequenced for a subset of the total (750 to ~20,000 taxa), plus 604 morphologic characters that applied across most of the data set. Nearly all (92%) of the cells in the resulting data matrix (73,060 taxa x 9535 characters) were empty due to lack of data. Nonetheless, the parsimony analysis recovered most eukaryotic groups down to the level of order as monophyletic taxa. The analysis utilized TNT software previously developed (and made publicly available) by Goloboff and colleagues and took 2.5 months on 3 desktop computers (total 96 GB RAM, 16 x 3 Ghz processors). To manage the flow of data, nearly all steps were automated from extracting, labeling, and aligning GenBank sequences to analyzing monophyly of groups at various taxonomic levels.
Looking ahead, the authors see biggest challenges not in tree-building, but in alignment software and “that the sequence information required is simply non-existent, and the morphological information is scanty and fragmentary.” I know that a short segment of a single mitochondrial gene is considered insufficient for phylogeny, but it would be interesting to see what TNT could do with 40,777 COI sequences from 6,506 fish species (FishBOL), for example. I imagine that even TNT might have trouble analyzing all 603,002 COI sequences of the 57,159 species represented in BOLD (with many more to come). Phylogenetic trees are established as the goal of evolutionary analysis, but we may need alternate methods for analyzing differences and similarities in very large data sets.
Hello:
I do not see the point in analyzing more than one sample of each species, given that sequenced ones were (I guess) identified by (true and competent) ichthyologists (in the case of fishes, obviously). And at this point, to analyze 6505 seqs in TNT is much easy if the sequences are aligned; as you noted, the main problem with such analyses is the alignment. Even to analyze the 40777 seqs is easier (or much easier depending on the structure of the data) than the analysis we did.
However, I was surfing a little the FishBol site, and I didn’t find the sequences… Are they publicly available?
Cheers; Marcos.
Dear Mark,
thanks for the post.
One of the things we discuss in the paper is that the missing
entries in our matrix make it run much slower –a lot of the
speed in TNT comes from figuring out data structure, which is
harder for so many missing entries. After this analysis which
just came out, I’ve tried the 218,348 prokaryotes Morgan Price
posted up in http://www.microbesonline.org/fasttree. Even if
there’s 2 1/2 times more taxa (when you delete the ca. 23,000
duplicates), completing a wagner tree plus TBR takes about 24 hs.,
even shorter than for our matrix –fewer characters (1287), and
fewer missing entries, thus providing a more structured data set.
While our matrix swaps at a speed of 130 x 10^6 rearrangs/sec,
the taller but narrower and better structed prokaryote matrix
swaps at about 2700 x 10^6 rearrangs/sec.
So, given that the barcoding data sets would make a narrower matrix,
with few or no missing entries, perhaps it would not be so hard to run
them with TNT. I would be interested in giving it a try, if I get an
aligned matrix. And I know that is a big “if”. Also, to interpret
results, the taxon names would have to be in the proper format
(see our paper for details) –visual inspection of the trees is
out of the question.
Just two small inaccuracies in your account: our analysis used about
2.5 months of processor time, but our small cluster had 16
processors; the analysis of each of the two matrices (the combined,
and the molecules alone) was completed in a couple weeks.
And second, none of our coauthors is german –I guess you meant Steve
Farris. He was born in the U.S. but has lived in Sweden for
more than 15 years. So, that would make “two swedes” on the team.
Pablo, thanks for your response. Increased speed TNT algorithm with more complete data sets (which would apply to DNA barcode data as you note) is helpful and important to know. Regarding format of COI barcode sequences in BOLD, at present the downloadable fasta files are aligned and have species names but not names for higher taxonomic levels, although your analysis makes it clear that would be a desirable upgrade. Thanks for other corrections–entry edited!
Marcos, regarding fish COI barcode sequences, those publicly available (hopefully many more to be released in near future) are accessible at BOLD homepage http://www.barcodinglife.org under “Public Projects.” Regarding multiple sequences for species, I see two reasons to include these. First, given the primary rationale for compiling barcodes is to enable DNA-based species identification, then testing the whole set to document those that form independent clusters (and to highlight those that do not) is important. Second, for any taxonomic group identification of some specimens (in fish, larval forms are particularly difficult for example) can be difficult even for experts, plus there are groups with taxonomic uncertainty as to species limits. Also as in any database particularly those with multiple submitters, clerical errors may occur. So comprehensive analysis that flags those sequences that do not form independent coherent groups with with other members of species is important recurrent process in curating database and improving species-level taxonomy.