In Cladistics 25 Sept 2007, Steven Trewick from Massey University, New Zealand applies mtDNA to help sort out endemic flightless grasshoppers in genus Sigaus, which are restricted to mountainous alpine habitat on New Zealand’s South Island. Here we might expect a complex pattern of diversification. These are small, terrestrial, flightless, presumably non-vagile (ie don’t travel far) animals in a deeply fragmented habitat. Their habitat lies in New Zealand’s central mountains, the Southern Alps, formed by a geologically recent uplift 5 to 2 million years ago. Like other organisms restricted to elevated mountain terrain, they are effectively living on “sky islands.” In this setting, we might expect a plethora of relatively young species with very narrow ranges, with difficulty determining which forms merit species-level status.
 Trewick focused on Sigaus australis species complex, which includes the apparently widely-distributed S. australis, and 5 sympatric or parapatric species with much narrower ranges (S. childi, S. obelisci, S. homerensis, and 2 undescribed species). Within this complex he analyzed 160 individuals collected at 26 locations (mostly S. australis (136 individuals) and 1-13 individuals for the more restricted species). For mtDNA analysis, an approximately 600 bp region of 12-16S and about 500 bp of 3′ COI (ie not overlapping COI barcode region!) were examined.
Trewick focused on Sigaus australis species complex, which includes the apparently widely-distributed S. australis, and 5 sympatric or parapatric species with much narrower ranges (S. childi, S. obelisci, S. homerensis, and 2 undescribed species). Within this complex he analyzed 160 individuals collected at 26 locations (mostly S. australis (136 individuals) and 1-13 individuals for the more restricted species). For mtDNA analysis, an approximately 600 bp region of 12-16S and about 500 bp of 3′ COI (ie not overlapping COI barcode region!) were examined.
Although the 3′ COI fragment analyzed in this grasshopper paper has been utilized in a number of invertebrate mtDNA studies, it is just one of many mtDNA targets that give essentially equivalent phylogenetic information (eg, in this study COI and 12S-16S gave same results). The hodgepodge of mtDNA regions analyzed in species-level animal work means that most data cannot be compared or combined. In my view, ALL animal mtDNA studies should include the standard COI barcode (defined relative to the mouse mitochondrial genome as the 648 bp region that starts at position 58 and stops at position 705; https://barcoding.si.edu/PDF/DWG_data_standards-Final.pdf), plus of course any other regions of interest. Standardization on the barcode region ensures long-term usefulness, both as a reference for identification and for comparisons across the diversity of animals. In addition to a defined genic target region, DNA barcode standards have other advantages, including that records are linked to voucher specimens and list primer sequences and include bidirectional trace files and quality scores.
In the present study single-strand conformation polymorphism (SSCP) of a 380 bp 12S fragment was used to screen for differences, and then individuals with different SSCP results were subjected to sequencing, so in the end just 40 of 160 Sigaus sp grasshoppers were sequenced for COI. This also means that there is voucher data in GenBank for just these 40 individuals. Continuing down the DNA barcode standard checklist, primer sequences are not easily accessible (there is a published reference for the primers, but access requires article purchase), it is not stated if bidirectional sequencing was done, and trace files and quality scores are not provided. I hope that future studies on New Zealand orthopterans will include the 5′ COI region and the remaining information, as I believe this will increase their long-term utility both as an identification reference and for comparisons across diversity of animal life (>520,00K individuals representing >50,000 species in BOLD so far). There is a big opportunity for grasshopper specialists to contribute–the BOLD taxonomy browser contains records for only 191 of the approximately 10,000 species in family Acrididae!
To skip to the conclusion, the sequence analysis gave an entirely different picture than existing morphologic taxonomy. 12S-16S and COI gave identical results: four well-supported geographically-structured clades within the widespread S. australis morphospecies, 3 of which had partly overlapping ranges. The 5 described or proposed species in the complex nested within these clusters, with shared or similar mtDNA haplotypes to S. australis from the same region.
The author concludes that the results show that “haplotype sharing and paraphyly essentially invalidate the DNA barcoding approach.” I disagree. To my reading, the most parsimonious explanation is that 1) morphologic taxonomy has overlooked deeply divergent genetic lineages, which likely represent different species, in S. australis for over 100 years, and 2) a number of morphologically distinctive forms have arisen very recently.
In support of the first point I note that in April 2008 report “Diversity and taxonomic status of some New Zealand grasshoppers” by the same author and Simon Morris, “Attention needs to be given to the spatial distribution of diversity within [S. australis complex]…Further morphological study may support the splitting of one or more of the groups indicated by phylogenetic analysis of mtDNA sequences.”
 Regarding point 2, genetic methods including DNA barcoding may not resolve very young species. For Sigaus sp. grasshoppers, nuclear sequence data will help sort out whether these are young species or the products of recent hybridization or introgression.
Regarding point 2, genetic methods including DNA barcoding may not resolve very young species. For Sigaus sp. grasshoppers, nuclear sequence data will help sort out whether these are young species or the products of recent hybridization or introgression.
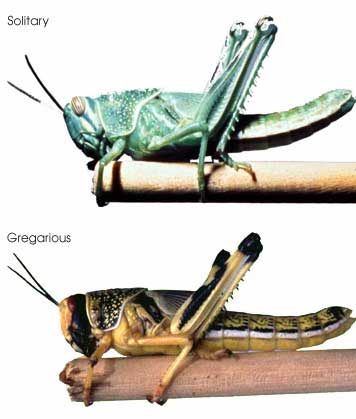 In this regard, I am struck by the apparent variability in some grasshopper species, as in the color morphs of S. childi shown above. It brings to my mind the extraordinary transformations from solitary grasshoppers to swarming locusts (these are members of the same Acrididae family as Sigaus). Perhaps grasshopper genetics include analogous latent “switches” that might enable relatively rapid evolutionary transformations.
In this regard, I am struck by the apparent variability in some grasshopper species, as in the color morphs of S. childi shown above. It brings to my mind the extraordinary transformations from solitary grasshoppers to swarming locusts (these are members of the same Acrididae family as Sigaus). Perhaps grasshopper genetics include analogous latent “switches” that might enable relatively rapid evolutionary transformations.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.