 The DNA barcode initiative aims to establish a universal identification system for plant and animal species by analyzing a standardized genetic locus (or for plants, a small set of loci). In addition to making analysis cheaper, standardizing on one or a few loci enables a diverse assemblage of researchers to work together to build an interoperative library.
The DNA barcode initiative aims to establish a universal identification system for plant and animal species by analyzing a standardized genetic locus (or for plants, a small set of loci). In addition to making analysis cheaper, standardizing on one or a few loci enables a diverse assemblage of researchers to work together to build an interoperative library.
If there were no Human Genome Project, researchers working gene by gene might eventually have decoded the human genome sometime during this century, albeit at much slower pace using more expensive and less accurate technology. For a genetic library of biodiversity, a concerted effort is essential. The various taxon-specific genetic initiatives, which are typically aimed at reconstructing deep evolutionary history, are too limited in scope (ie number of species and individuals per species analyzed) and too expensive in terms of cost per species to completely catalog animal and plant life. In addition, because different groups analyze different gene regions, it is impossible to stitch together the results into single database, for instance one that could be used to identify an unknown specimen without knowing beforehand what group it belongs to. The DNA barcoding initiative offers the necessary framework for constructing a genetic reference database for species. In addition as a large-scale project it should help drive technological improvements analogous to those spawned by the Human Genome Project which enabled its completion for a fraction of the originally projected cost.
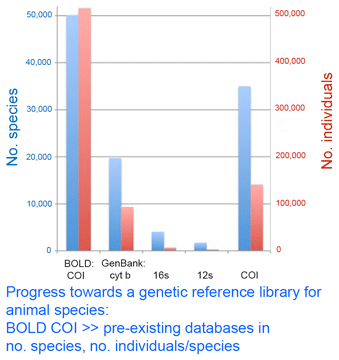
As of today, researchers have deposited 516,134 barcode records from 50,138 species in Barcode of Life Database (BOLD) www.barcodinglife.org. According to my analysis of GenBank shown in figure, this puts COI BOLD records far above the totals for any other single gene for animals. Thus five years of a concerted, standardized approach has leapt ahead of 30 years of incremental analysis. If the proof is in the pudding, this to me is a pudding that proves the value of the DNA barcoding initiative. Comparison of the totals indicates that most BOLD COI records are not yet in GenBank, although some aspects are visible through ID engine and Taxonomy Browser, so there is work to help move these fully into the public domain and at the same time ensure appropriate academic credit. Congratulations to all those moving this effort forward.
Very impressive indeed. I agree about the value of getting these into GenBank. Although BOLD is quite powerful in some ways the value of these records is reduced by not being open access. The excellent Hebert et al 2003 paper downloaded and analysed all available COI sequences, I don’t see it possible to do anything similar anymore given that most sequences are now restricted. Some creative thought needed as you say to change this while maintaining academic credit. Interesting times ahead for large scale analyses (I hope).
—
Hebert et al. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci (2003) vol. 270 Suppl 1 pp. S96-9