Visualizing birds so far
 In 2004 PLoS Biology, Hebert and colleagues (I am a co-author) observed that differences in COI barcodes among 260 species of North American birds were generally much larger than those within species, with the result that “distinguishing species was generally straightforward.” In addition, we noted 4 birds with large intraspecific divergences that likely represented overlooked species. Our study included only about 1/50 of world birds (out of approximately 10,000 named species) and modest sampling of differences within species (multiple individuals (average 2.4, range 2-10) for 130 species), so not surprisingly some scientists wondered about the generalizability of the findings in birds in particular and animals in general. In an accompanying commentary, Cicero and Moritz wrote “…a true test of the precision of mtDNA barcodes to assign individuals to species…would require that all members of a genus be examined, rather than a random sample of imprecisely-defined close relatives, and that taxa be included from more than one geographic region.” They concluded their essay:
In 2004 PLoS Biology, Hebert and colleagues (I am a co-author) observed that differences in COI barcodes among 260 species of North American birds were generally much larger than those within species, with the result that “distinguishing species was generally straightforward.” In addition, we noted 4 birds with large intraspecific divergences that likely represented overlooked species. Our study included only about 1/50 of world birds (out of approximately 10,000 named species) and modest sampling of differences within species (multiple individuals (average 2.4, range 2-10) for 130 species), so not surprisingly some scientists wondered about the generalizability of the findings in birds in particular and animals in general. In an accompanying commentary, Cicero and Moritz wrote “…a true test of the precision of mtDNA barcodes to assign individuals to species…would require that all members of a genus be examined, rather than a random sample of imprecisely-defined close relatives, and that taxa be included from more than one geographic region.” They concluded their essay:
“But to determine when and where this approach [i.e., DNA barcoding] is applicable, we now need to discover the boundary conditions. The real challenge lies with tropical taxa and those with limited dispersal and thus substantial phylogeographic structure. Such analyses need to be taxonomically broad and need to extend beyond the focal geographic region to ensure that potential sister taxa are evaluated and can be discriminated. There is also the need to examine groups with frequent (possibly cryptic) hybridization, recent radiations, and high rates of gene transfer from mtDNA to the nucleus.”
As of today, the BOLD taxonomy browser at Phylum Chordata, Class Aves (www.boldsystems.org/views/taxbrowser.php?taxid=51 indicates over 24,000 barcoded avian specimens representing over 3,800 avian species, nearing 40% of world avifauna. By my count there are over 30 publications on DNA barcoding in birds, including large surveys in North America, Scandinavia, Argentina, Brazil, and Korea.
In the next few posts, I try to look at what we have learned so far, with an emphasis on visual representation. The short answer to the technical question of barcoding effectiveness in birds is that the early observations are borne out, with a few interesting exceptions. My rough summary is that about 95% of bird species can be distinguished by DNA barcode, the remainder are sorted into pairs or small sets of closely-related species, and about 10% of named species show large divergences that likely represent unrecognized species. This last observation brings up an important point–taxonomy is undergoing constant revision, even in a group as well-studied as birds. For example, over the past 30 years, about 10% of the roughly 2000 bird species on the American Ornithologists’ Union Check-list have had species limits revised, and this process is not near closure. So when we compare to sequence data to taxonomic classification, we have to keep in mind the latter is a moving target.
Looked at more broadly, the central finding is that mtDNA sequence differences in birds partition into distinct clusters. Most mtDNA sequence clusters correspond to a single named species, and the ongoing process of taxonomic revision is tightening the one-to-one correspondence between clusters and species designations. In fact, a person with no knowledge of avian biology could closely approximate species numbers and limits simply by sorting COI barcodes into sequence clusters. Of course, the concordance of species limits and mtDNA sequence differences is not a new observation (see for example Avise et al 1987, 1999; Moore 1995), but it is now backed up by much more data. An important but unsolved question is why mtDNA partitions into narrow clusters in birds and other animals. One or more of the proposed mechanisms may turn out to be correct but none has been proven so far.
To begin visualization survey:
Pairwise sequence differences within most bird species are small, usually much less than 1%. (“Pairwise sequence differences” means comparing each individual to every other individual of the same species; for n individuals there are n(n-1)/2 comparisons.) In looking at this data, I think that the absolute scale is important. One of the great benefits of working with a standard barcode region is that we can compare results across diverse taxa. To get into particulars, genetic differences are roughly similar in mtDNA protein coding genes (e.g. COI, cytb), but divergences in the mitochondrial control region are an order of magnitude greater.
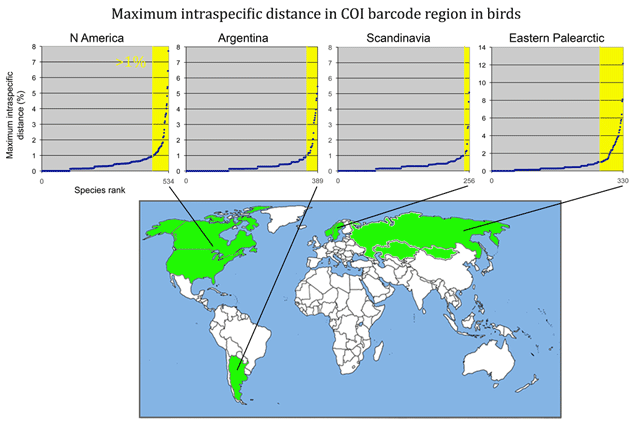
Here is a look at MAXIMUM intraspecific distances (K2P metric) among some of the larger geographically-based surveys published so far. (Most of this information can also be found in published papers cited below.) The aim is to see what we can learn from outliers to the general observation of narrow differences within species. For these illustrations, I went to the Public Projects section of BOLD www.barcodinglife.org, selected a project (Birds of North America Phase II, Kerr 2007; Birds of Argentina Phase I, Kerr et al 2009a; Birds of Scandinavia, Johnsen et al 2010; Birds of the eastern Palearctic, Kerr et al 2009b) and ran a “Nearest Neighbor” analysis with BOLD software, which calculates average and maximum intraspecific distance, as well as identity of and distance to the “nearest neighbor”. The results were copied and pasted into an Excel spreadsheet, sorted by increasing maximum intraspecific distance, and displayed in a graph as shown below (note different y-axis scale for eastern Palearctic). The total number of species are noted on the x-axis; yellow marks those with >1% maximum intraspecific distance. The curves are roughly similar among the regions except that the proportion >1% differs.
In the next post, I look more carefully at the apparent outliers. What makes them different–biology or taxonomy?
Note added 23 March 2011: Kevin Kerr points out that “eastern Palearctic” refers to entire region east of Europe, and thus the eastern Palearctic survey referred to above includes sites spanning most of Russia, Kazakhstan, and Mongolia (not just the eastern half of Russia as highlighted in map).
Note added 24 March 2011: Map corrected to show collecting region for eastern Palearctic survey.
This entry was posted on Monday, March 21st, 2011 at 9:59 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.