Ground beetles join in
 On September 25, 2010, BOLD passed 1 M barcode records, and the International Barcode of Life ( iBOL) was officially launched in Toronto, Canada, with a goal of 5 M records representing 500 K species in 5 years, the largest biodiversity genomics project to date. In terms of DNA sequencing, the iBOL targets (5 x 106 barcodes x 650 bp/barcode = 3.3 x 109 bp) are equivalent to the Human Genome Project (human genome = 3.4 x 109 bp). However, whereas HGP involved sequencing DNA samples from a few individuals, the DNA barcode library is built by thousands of scientists examining thousands of individual specimens, one by one. So a big challenge is obtaining, identifying, tracking, processing, and preserving millions of specimens.
On September 25, 2010, BOLD passed 1 M barcode records, and the International Barcode of Life ( iBOL) was officially launched in Toronto, Canada, with a goal of 5 M records representing 500 K species in 5 years, the largest biodiversity genomics project to date. In terms of DNA sequencing, the iBOL targets (5 x 106 barcodes x 650 bp/barcode = 3.3 x 109 bp) are equivalent to the Human Genome Project (human genome = 3.4 x 109 bp). However, whereas HGP involved sequencing DNA samples from a few individuals, the DNA barcode library is built by thousands of scientists examining thousands of individual specimens, one by one. So a big challenge is obtaining, identifying, tracking, processing, and preserving millions of specimens.
 What are recent arrivals to library? For one example, in current Frontiers Zool, researchers from Germany and US (I am co-author) report on DNA identification of Central European ground beetles (family Carabidae). This family comprises “no less than an estimated 40,000 described species that inhabit all terrestrial habitat types from the sub-arctic to wet tropical regions,” making identifications a challenge for taxonomists and non-specialists alike. Raupach and colleagues successfully amplified and sequenced COI barcodes and nuclear ribosomal DNA expansion segments D3, V4, and V7, from 344 specimens representing 75 species in 28 genera (average 4 specimens/species, range 2-13). Most specimens were preserved in 96% alcohol for 1-2 years; some were stored as dry pinned specimens for up to 12 years. 73 (97%) species were resolved by COI, whereas the 3 nuclear markers individually resolved a smaller proportion, 81% (D3), 57% (V4) and 87% (V7), and combining the 3 nuclear markers gave 95% discrimination. The one species pair with shared COI haplotypes also showed identical nuclear markers. Two species exhibited distinct COI clusters (intra-specific p-distances 2.7%, 3.8%), 1 of which also had distinct nuclear haplotypes.
What are recent arrivals to library? For one example, in current Frontiers Zool, researchers from Germany and US (I am co-author) report on DNA identification of Central European ground beetles (family Carabidae). This family comprises “no less than an estimated 40,000 described species that inhabit all terrestrial habitat types from the sub-arctic to wet tropical regions,” making identifications a challenge for taxonomists and non-specialists alike. Raupach and colleagues successfully amplified and sequenced COI barcodes and nuclear ribosomal DNA expansion segments D3, V4, and V7, from 344 specimens representing 75 species in 28 genera (average 4 specimens/species, range 2-13). Most specimens were preserved in 96% alcohol for 1-2 years; some were stored as dry pinned specimens for up to 12 years. 73 (97%) species were resolved by COI, whereas the 3 nuclear markers individually resolved a smaller proportion, 81% (D3), 57% (V4) and 87% (V7), and combining the 3 nuclear markers gave 95% discrimination. The one species pair with shared COI haplotypes also showed identical nuclear markers. Two species exhibited distinct COI clusters (intra-specific p-distances 2.7%, 3.8%), 1 of which also had distinct nuclear haplotypes.
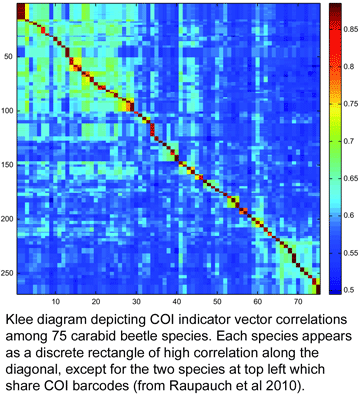 To my knowledge, this is the first taxonomic paper with a “Klee diagram” depicting indicator vector correlations among COI barcode sequences. As developed by mathematician Larry Sirovich and his colleague Yu Zhang (Sirovich et al PloS ONE 2010), indicator vectors are digital representations of DNA sequences that “preserve diagnostic character probabilities, enable automated classification of test sequences, and generate high-information density single-page displays” such as Klee diagram shown here. According to BOLD Taxonomy Browser, there are DNA barcodes for 495 carabid beetle species so far, so I look forward more of the remaining 39,505 or so species joining the barcode library, and dream of a comprehensive indicator vector/Klee analysis of ground beetle family.
To my knowledge, this is the first taxonomic paper with a “Klee diagram” depicting indicator vector correlations among COI barcode sequences. As developed by mathematician Larry Sirovich and his colleague Yu Zhang (Sirovich et al PloS ONE 2010), indicator vectors are digital representations of DNA sequences that “preserve diagnostic character probabilities, enable automated classification of test sequences, and generate high-information density single-page displays” such as Klee diagram shown here. According to BOLD Taxonomy Browser, there are DNA barcodes for 495 carabid beetle species so far, so I look forward more of the remaining 39,505 or so species joining the barcode library, and dream of a comprehensive indicator vector/Klee analysis of ground beetle family.
In closing, professional and non-professional insect specialists alike may may enjoy recently released film “Beetle Queen Conquers Tokyo” by Jessica Oreck, a lyrical look at beetle and insect fanciers in Japan.
This entry was posted on Monday, September 27th, 2010 at 5:08 pm and is filed under General. You can follow any responses to this entry through the RSS 2.0 feed. Both comments and pings are currently closed.